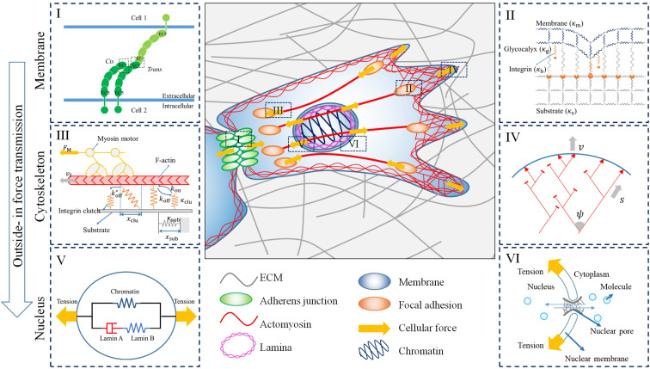
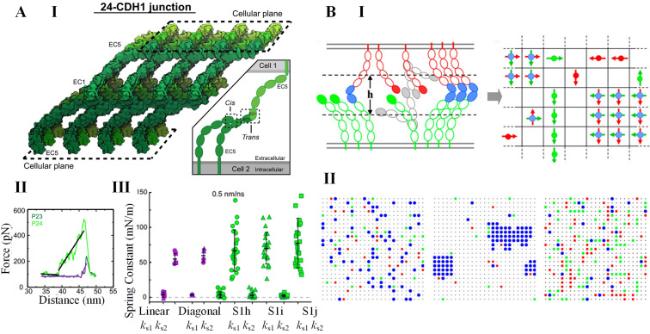
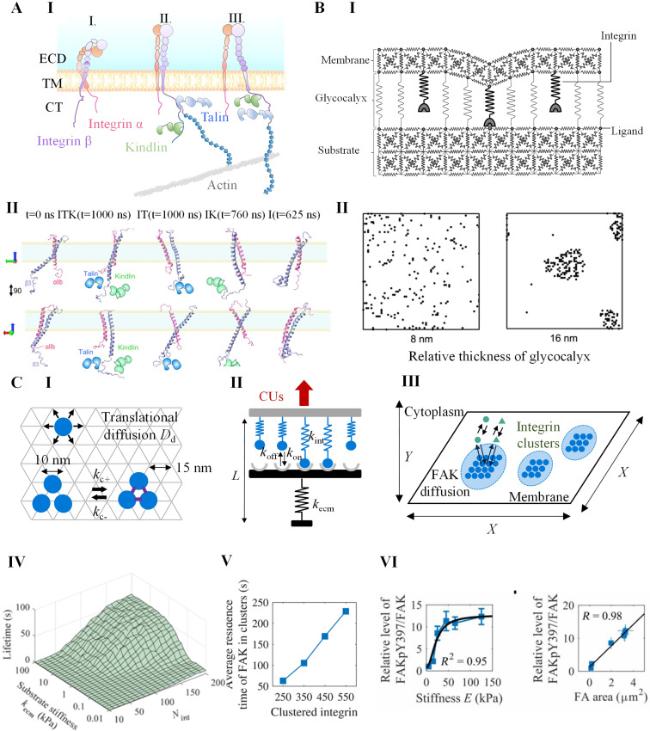
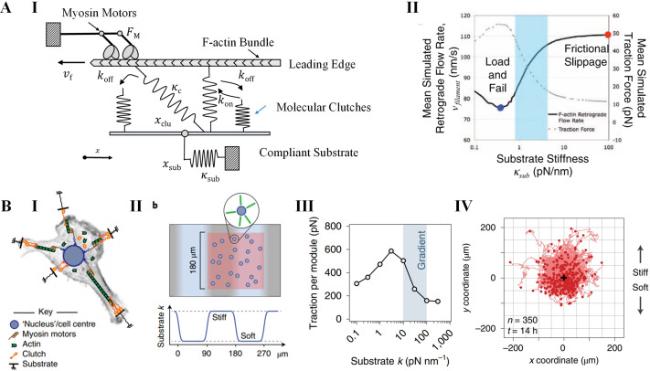
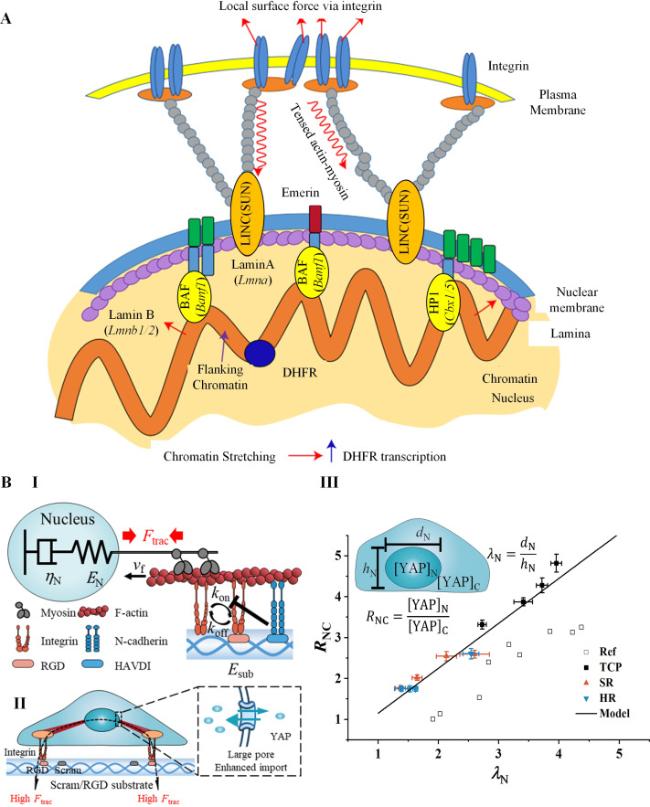
Cell-cell interactions are primarily established through cadherin-mediated adherens junctions, which not only bind cells together but also function as mechanotransducers [
33,
34,
35,
36]. Adherens junctions can be classified into three types based on their shapes and stability: (i) zonula adherens, (ii) punctum adherens, and (iii) multicellular adherens [
37]. The stability of adherens junctions is maintained by calcium-dependent adhesion molecules called cadherins [
38]. Cadherin is a superfamily that consists of various members, including classical cadherins, desmosomal cadherins, and protocadherins (Pcdhs) [
39]. Classical cadherins have five extracellular repeats (EC1-EC5), a transmembrane domain, and an intracellular domain [
40]. Based on their ectodomain sequence, classical cadherins can be further divided into two subfamilies: (i) type I, which includes E-cadherin and N-cadherin, characterized by a HAV peptide motif and a conserved tryptophan residue (Trp2), and (ii) type II, which includes VE-cadherin, characterized by two conserved tryptophan residues (Trp2 and Trp4) but lacking the HAV motif [
41]. These classical cadherins form two types of interactions. The first is
trans interaction, which connects cadherins on two adjacent cells, and the second is
cis interaction, which connects cadherins on the same cell [
33].
Trans interactions, facilitated by a tryptophan exchange between two EC1 domains, can adopt two distinct conformations: strand-swap (S-dimer) and X-dimer [
42]. The lower-affinity X-dimers preferentially transform into the more stable S-dimers, and this transformation process is dependent on actomyosin-generated tensile forces [
43,
44]. On the other hand,
cis interactions aid in cadherin clustering through both extracellular and intracellular domains [
33]. Extracellular
cis interactions (EC-
cis interactions) occur between the EC1 domain and EC2-3 domains of classical cadherins [
45]. However, cadherin clusters formed solely based on EC-
cis interactions are small and unstable [
33]. The instability of EC-
cis interactions can be significantly reinforced by the formation of an intracellular cadherin-catenin complex (CCC), which relies on
trans binding and links the clusters to actin filaments [
46,
47].