1. Introduction
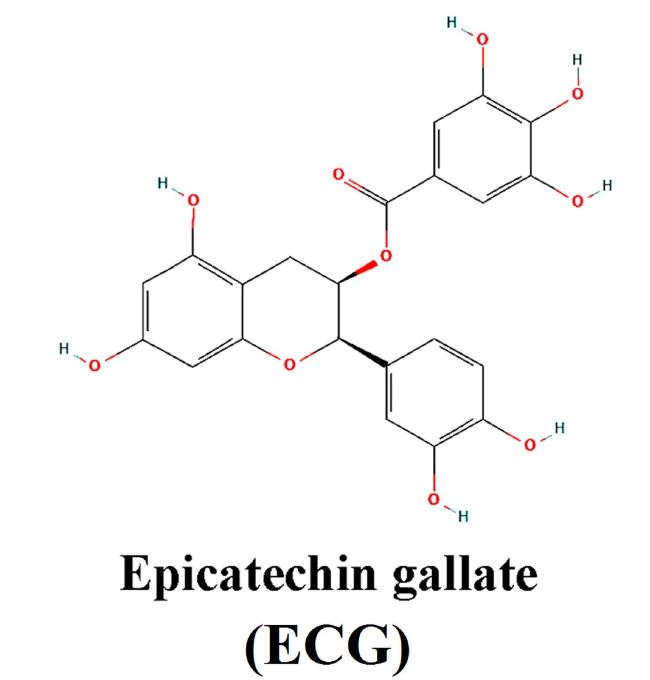
Fig. 1. Chemical structure of ECG. |
2. Materials and methods
2.1. Structure retrieval and preparation of ligand
2.2. Structure retrieval and preparation of SARS-CoV-2 proteins and entry factors
2.3. Docking procedure
3. Results
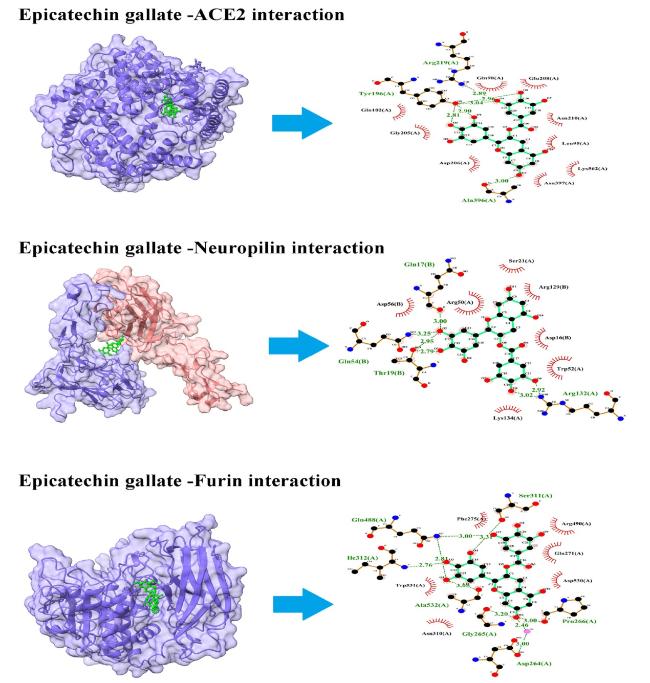
3.1. Binding of ECG with SARS-CoV-2 entry factors
3.1.1. ACE2
3.1.2. Neuropilin
3.1.3. Furin
Fig. 2. Interaction of ECG with entry factors. ECG blocks several amino acid residues of ACE2, neuropilin, and furin at binding pockets via H- bonds and other non-covalent interactions. Left panel shows the site of protein-ligand interactions. Right panel depicts the amino acid residues of concerned protein involved in H-bonds and hydrophobic interactions with ECG. |
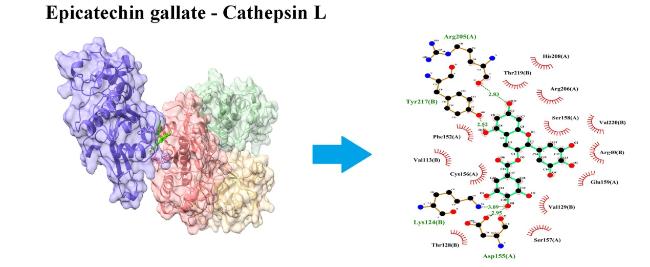
3.1.4. CTSL
Fig. 3. Interaction of ECG with cathepsin L (CTSL). ECG blocks several amino acid residues of CTSL at binding pockets via H- bonds and other non-covalent interactions. Left panel shows the site of protein-ligand interactions. Right panel depicts the amino acid residues of CTSL involved in H-bonds and hydrophobic interactions with ECG. |
3.2. Binding of ECG with SARS-CoV-2 proteins
3.2.1. Spike protein RBD
3.2.2. Main protease (Mpro)/NSP5
3.2.3. Papain-like protease (PLpro)
Fig. 4. Interaction of ECG with viral spike protein and proteases. ECG blocks several amino acid residues of receptor binding domain (RBD) of spike protein, main protease (Mpro), and papain like protease (PLpro) at binding pockets via H- bonds and other non-covalent interactions. Left panel indicates the site of protein-ligand interactions. Right panel represents the amino acid residues of concerned protein involved in H-bonds and hydrophobic interactions with ECG. |
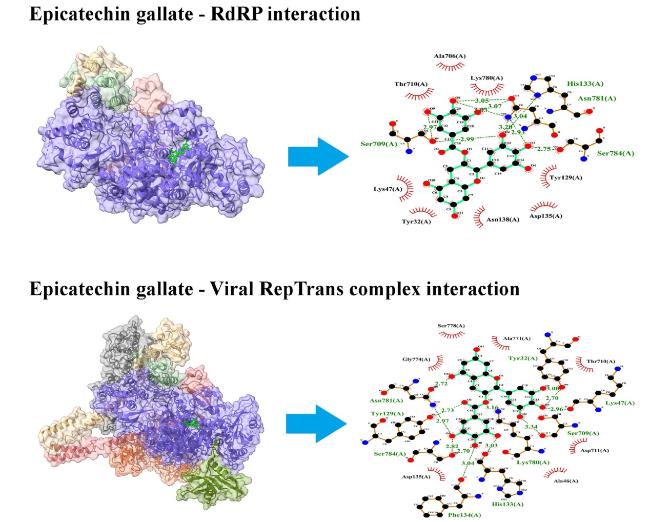
3.2.4. RNA-dependent RNA polymerase (RdRP)/NSP12
3.2.5. RepTrans complex
Fig. 5. Interaction of ECG with viral RdRP and RepTrans complex. ECG blocks several amino acid residues of RdRP and RepTrans complex at binding pockets via H-bonds and non-covalent interactions. Left panel shows the site of protein-ligand interactions. Right panel depicts the amino acid residues of concerned protein involved in H-bonds and hydrophobic interactions with ECG. |
3.2.6. NSP7-NSP8 complex
3.2.7. NSP13
Fig. 6. Interaction of ECG with viral NSP7-NSP8 complex and NSP13. ECG blocks several amino acid residues of NSP7-NSP8 complex and NSP13 at binding pockets via H-bonds and other non-covalent interactions. In the left panel, sites of protein-ligand binding have been presented. Amino acid residues of concerned protein involved in H-bonds and hydrophobic interactions with ECG have been presented in right panel. |
3.2.8. NSP14
3.2.9. NSP15
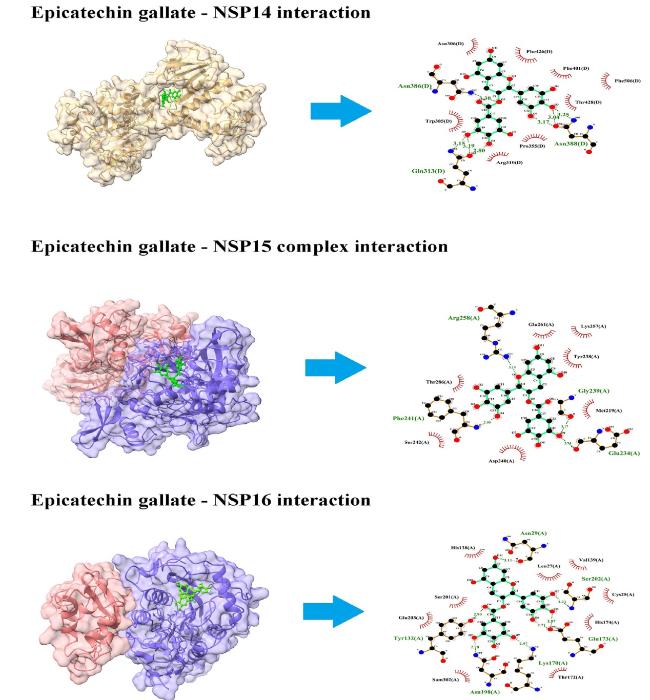
3.2.10. NSP16
Fig. 7. Interaction of ECG with viral NSP14, NSP15, and NSP16. ECG blocks several amino acid residues of NSP14, NSP15, and NSP16 at binding pockets via H-bonds and non-covalent interactions. Left panel shows the site of protein-ligand interactions. Right panel depicts the amino acid residues of concerned protein involved in H-bonds and hydrophobic interactions with ECG. |
4. Discussion
Table 1. Binding affinity of protein-ligand complex. |
| Protein ligand complex | Binding affinity (kcal/mol) |
|---|---|
| ECG ∼ ACE-2 | −8.3 |
| ECG ∼ Neuropilin | −8.0 |
| ECG ∼ Furin | −8.9 |
| ECG ∼ Cathepsin L | −10.2 |
| ECG ∼ spike protein RBD | −7.4 |
| ECG ∼ COVID-19 Mpro/NSP5 | −8.4 |
| ECG ∼ PLpro | −8.9 |
| ECG ∼ RdRP/NSP12 | −8.6 |
| ECG ∼ RepTrans complex | −9.0 |
| ECG ∼ NSP7-NSP8 complex | −8.3 |
| ECG ∼ NSP13 | −8.7 |
| ECG ∼ NSP14 | −9.1 |
| ECG ∼ NSP15 | −7.9 |
| ECG ∼ NSP16 | −8.5 |
Table 2. Hydrogen bonds and Hydrophobic interactions established between ECG and amino acid residues of host entry factors/viral proteins of SARS-CoV-2. |
| Protein ligand complex | Amino acids of proteins establishing Hydrogen bonds with ligand | Amino acids of proteins establishing hydrophobic interactions with ligand |
|---|---|---|
| ECG ∼ ACE-2 | Tyr196; Arg219; Ala396 | Gln102; Gly205, Gln98, Asp206; Glu208, Asn210, Leu95, Lys562, Asn397 |
| ECG ∼ Neuropilin | Gln17; Gln54, Thr19, Arg132 | Asp56, Arg50, Ser21, Arg129, Asp16, Trp52, Lys134 |
| ECG ∼ Furin | Ile312; Gln488; Ser311; Ala532, Asp264, Pro266; Pro266 | Phe275; Glu271; Arg490; Asp530; Asn310; Trp531 |
| ECG ∼ Cathepsin L | Lys124; Asp155; Arg205; Tyr217 | Val113(B); Phe152(A); Cys152(A); Thr219(B); His208(A); Arg206(A); Aer158(A); Arg40(B); Val220(B); Glu159(A); Val129(B); Ser157(A); Thr128(B); Cys156(A) |
| ECG ∼ spike protein RBD | Asp415; Leu503; Phe501; Met417; Pro450 | Glu502; Ser500; Phe451; Pro413 |
| ECG ∼ Mpro/NSP5 | His41; Cys145; His163; Phe140; Thr190 | Leu141; Glu166; Met165; Gln189; Pro168 |
| ECG ∼ PLpro | Asp108; Asn109; Thr158; Glu161; Val159; Gln269 | Glu161; Leu162; Asn109; Val159; Gly160; Leu162; Gln269; Gln269 |
| ECG ∼ RdRP/NSP12 | His133; Asn781; Ser784; Ser709 | Thr710; Ala706; Lys780; Tyr129; Asp135; Asn138; Tyr32; Lys47 |
| ECG ∼ RepTrans complex | Asn781; Ser784; Tyr129; Ser784; Tyr32; Lys47; Ser709; Lys780; His133; Phe134 | Gly774; Aer778; Ala771; Thr710; Asp711; Ala46; Asp135 |
| ECG ∼ NSP7-NSP8 complex | Ser15; Gln18; Glu19 | Met87; Arg21; Gln18; Leu14; Arg80; His36; Asn37; Val83 |
| ECG ∼ NSP13 | Ala4; Glu136; Leu235; Arg129 | Arg21(A); Pro23(A); Phe24(A); Gly3(A); Leu32(A); Val6(A); Pro234(A); Phe133(A) |
| ECG ∼ NSP14 | Gln313; Asn386; Asn388 | Trp385; Asn306; Phe426; Phe401; Phe506; Thr428; Pro355; Arg310 |
| ECG ∼ NSP15 | Glu234; Phe241; Gly239; Arg258 | Met219; Tyr238; Asp240; Ser242; Thr286; Glu261; Lys257 |
| ECG ∼ NSP16 | Tyr132; Asn29; Asn198; Lys170; Glu173; Ser202 | Glu203; Ser201; His138; Leu27; Val139; Cys25; His174; Thr172 |
Fig. 8. Multi-target potency of ECG for SARS-CoV-2 entry and replication within the host cell. Spike protein of SARS-CoV-2 attaches with host ACE2 and undergoes proteolytic cleavage by host proteases viz. furin and neuropilin. This facilitates fusion and viral entry into the host cell. Cathepsin L (CTSL) helps in release of (+) ss viral RNA to the host cytoplasm. Viral genome uses host-translational machinery to generate viral polyproteins (pp1a, pp1b). pp1a generates viral proteases viz. Mpro and PLpro that cleaves pp1b into several non-structural proteins (NSPs). NSPs constitute the replication-transcription machinery for replication of viral genome and synthesis of (−) ss viral RNA is crucial for synthesis of structural proteins and their packing into new virions. These virions are then exocytosed out of the cells to infect the fresh one. ECG can bind with host entry factors, viral proteases, and NSPs to inhibit viral entry and replication within the host cell. |