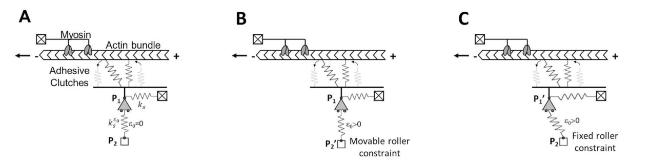
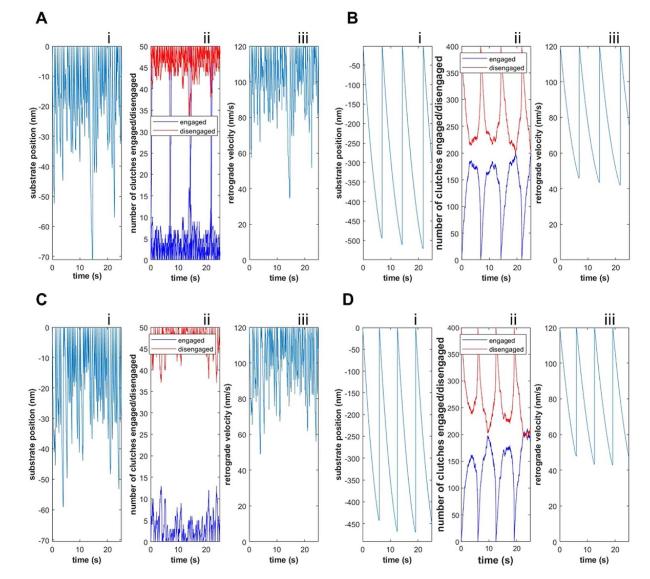
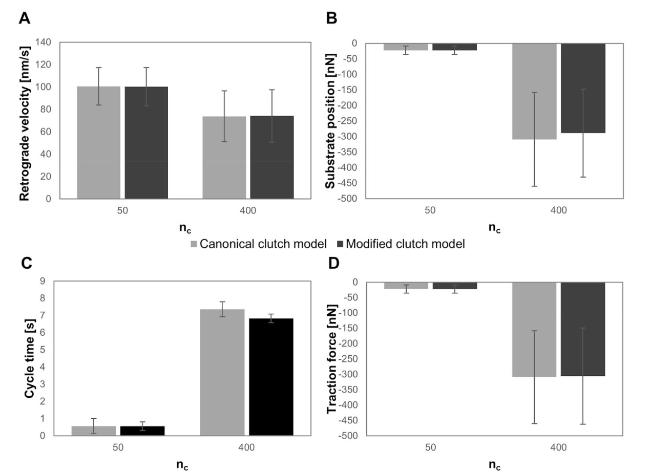
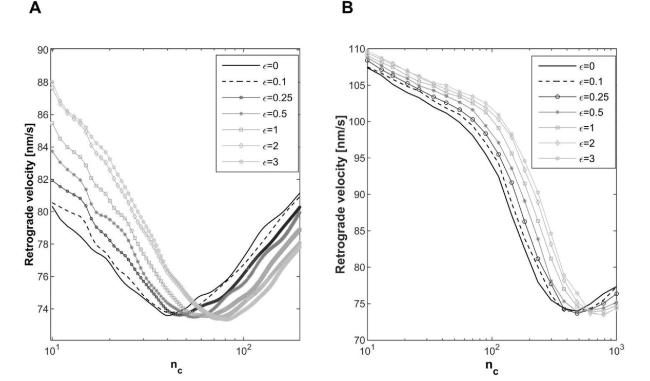
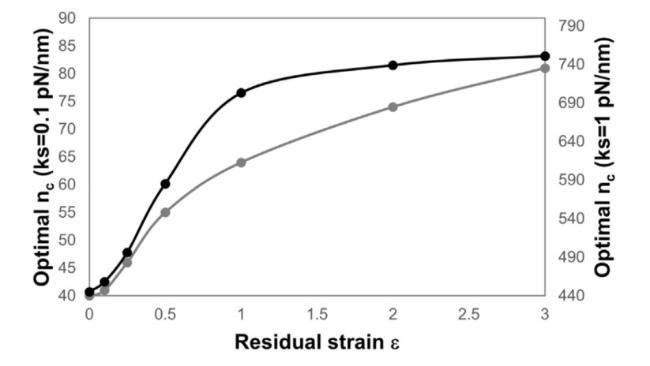
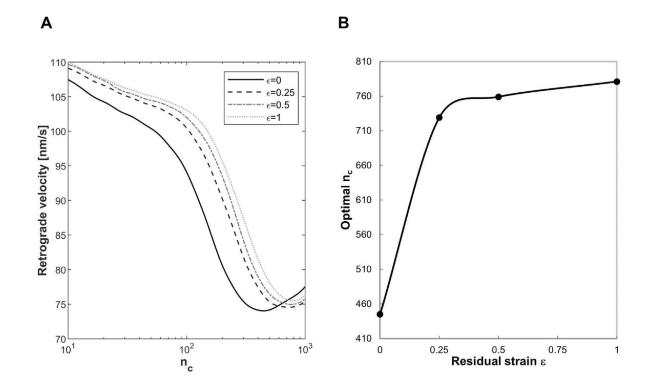
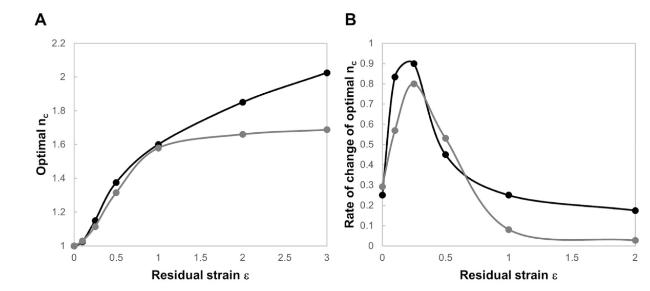
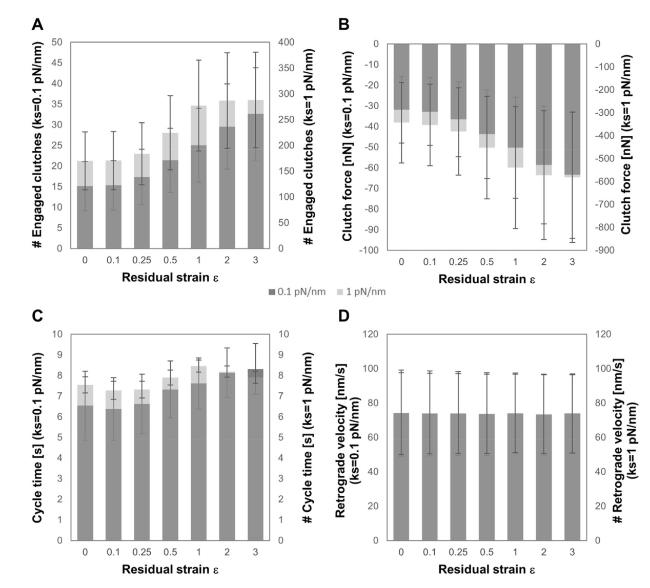
Cells in living tissues are continuously exposed to a plethora of biophysical stimuli coming from their microenvironment, i.e., cells [
1,
2,
3] and the extracellular matrix (ECM) [
4,
5,
6]. A growing body of evidence indicates the importance of the mechanical microenvironment in modulating and guiding cell behaviour in both physiological and pathological conditions [
7,
8,
9]. In this context, ECM mechanics has emerged as a key regulator of cellular fate. Indeed, strong evidence indicates that ECM elasticity plays an important role in regulating cell adhesion and cytoskeleton organisation that ultimately affect proliferation, migration, self-renewal and differentiation [
8,
10,
11,
12]. It is important to note that most of the research exploring the role of ECM elasticity on cell behaviour, has been performed in 2D contexts. The comprehension of its importance in three dimensions seems challenging since it is difficult to independently control matrix stiffness and mesh-size, two parameters that, in 3D matrix, affect cell adhesion and spreading in opposite manner [
13]. However, Yuan et al. have recently reported that a local stiffening of 3D hydrogel network produces adhesion, spreading and osteogenic differentiation of stem cells as observed on 2D stiff substrates [
10,
14]. In recent years, numerous works have also highlighted the important role of time dependent mechanical features of the microenvironment, i.e., viscoelasticity, on cell mechanosensing and mechanotransduction processes. ECM viscoelastic moduli have proved to be effective in regulating cell adhesion, spreading, proliferation, and differentiation [
15,
16,
17,
18,
19,
20]. The role of the ECM dynamic behaviour was also investigated by considering the effect of ligand mobility (i.e., surface viscosity) on cell adhesion and spreading [
21]. Beside ECM elastic and viscoelastic moduli, also the strain energy accumulated within the ECM plays an important role in cell and tissues physiological and pathological conditions. For instance, residual stresses, defined as stresses persisting in the tissue solid phase when all the external loads are removed, have been demonstrated as key regulators in physiology of tissues as veins and arteries [
22,
23,
24,
25] as well as in solid tumours [
26,
27,
28,
29]. Residual stresses, generally originated by a non-isotropic tissue growth and/or remodelling [
30,
31], are important features that together with ECM elastic and viscoelastic moduli uniquely define the mechanical identity of cellular microenvironment. However, most of the literature studies aimed at elucidating the basic mechanism underlying the mechanoregulation in tissue morphogenesis and histogenesis are limited to the role of elastic modulus of the ECM, neglecting or downplaying the important role of residual stresses.