Introduction
Fig. 1 Classification of ncRNAs. Housekeeping RNAs have been extensively studied because of the fundamental nature of their expression. Other ncRNAs can be synthesized because of external stimuli and may be classified according to length into small RNAs (20-24 nucleotides) and long non-coding RNAs (≥200 nucleotides) |
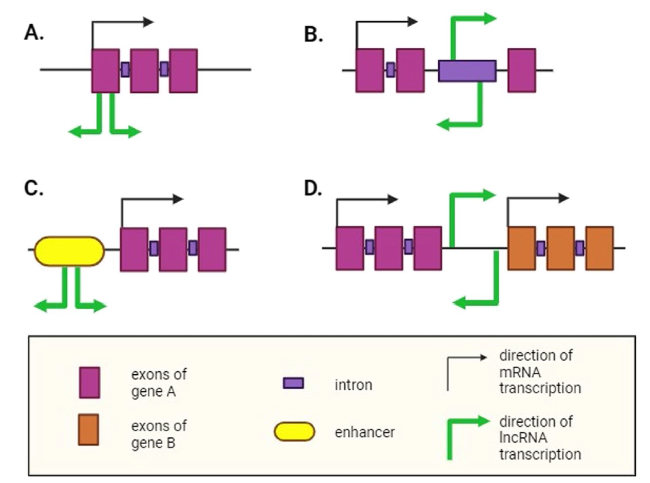
Fig. 2 Classification of lncRNAs. Based on the genomic locations from which RNAs are transcribed relative to protein-coding regions, long non-coding RNAs (lncRNAs) can be classified into five distinct groups. A Exonic lncRNAs can partially or completely overlap with the exons of genes that code for proteins. They are transcribed in either the same direction (sense lncRNA) or the opposite direction (antisense lncRNA) as the mRNA. B Intronic lncRNAs originate from long introns of protein-coding genes and may be transcribed in either the same or the opposite direction as the mRNA. C Enhancer lncRNAs are transcribed from short enhancer regions of DNA. They may be bidirectional (as shown in the figure) or unidirectional, depending on their transcription direction. D Long intergenic non-coding RNAs (lincRNAs) arise from the intergenic region between two protein-coding genes and may be transcribed in either the same or the opposite direction of the nearest genes |
Functional mechanisms of lncRNAs
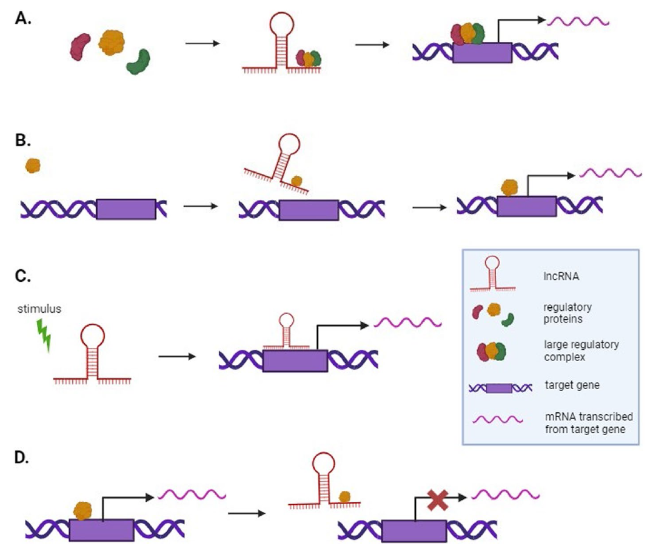
Fig. 3 Mechanisms of action of lncRNAs. LncRNAs may regulate gene expression by acting as scaffolds, guides, signals, or decoys. A Scaffolds bind to multiple small molecular components simultaneously and act as a platform for assembling these components into large regulatory complexes that activate or inhibit gene expression. B Guides bind to specific regulatory proteins and direct their localization to specific target genomic loci where gene expression is regulated. C Signals function as molecular cues because they precisely start transcription at certain times and places, letting cells respond to different stimuli. They function in various ways, such as by directly binding to the target site to perform a regulatory role. D Decoys act by binding to RNA-binding proteins, such as transcription factors or chromatin modifiers, and sequester them away from their intended targets, thus inhibiting these proteins from performing their roles. Note: The regulatory proteins are shown as positively regulating gene expression, but they may also inhibit gene expression |
Scaffolds
Guides
Signals
Decoys
Stress tolerance responses in plants
Table 1 Recent studies on lncRNAs in relation to plant stress (2019-2024) |
| Plant Species | No. of Differentially Expressed LncRNAs | Stress | Reference | |
|---|---|---|---|---|
| 1 | Capsicum annuum (bell pepper) | 1887 | osmotic | (Baruah et al. 2021) |
| 2069 | salt | |||
| 2101 | cold | |||
| 2833 | heat | |||
| 2 | Triticum aestivum (bread wheat) | 1515 | drought | (Li et al. 2022) |
| 3 | Camellia sinensis (tea) | 172 | salt | (Wan et al. 2020) |
| 4 | Brassica juncea (Indian mustard) | 1614 | heat and drought | (Bhatia et al. 2020) |
| 5 | Manihot esculenta (cassava) | 117 | drought | (Ding et al. 2019) |
| 6 | Melilotus albus (honey clover) | 550 | salt | (Zong et al. 2021) |
| 7 | B. napus (rapeseed) Q2 genotype | 126 | drought | (Tan et al. 2020) |
| B. napus (rapeseed) Qinyou 8 genotype | 359 | |||
| 8 | B. rapa (Chinese cabbage) | 93 | heat | (Eom et al. 2021) |
| 9 | Oryza sativa (rice) ssp. japonica | 97 | heat | (Zhang et al. 2021) |
| O. sativa (rice) ssp. indica | 103 | |||
| 10 | Panicum virgatum (switchgrass) | 368 | drought | (Guan et al. 2024) |
| 11 | Olea europaea (olive) | 2076 | heavy metal (Al) | (Wu et al. 2023) |
| 12 | Hordeum vulgare (barley) | 195 | heavy metal (Cd) | (Zhou et al. 2023) |
| 13 | Populus trichocarpa (black cottonwood) | 1183 | salt | (Ye et al. 2022) |
| 14 | Solanum pennellii (wild tomato) | 137 | salt | (Li et al. 2022) |
Salt stress
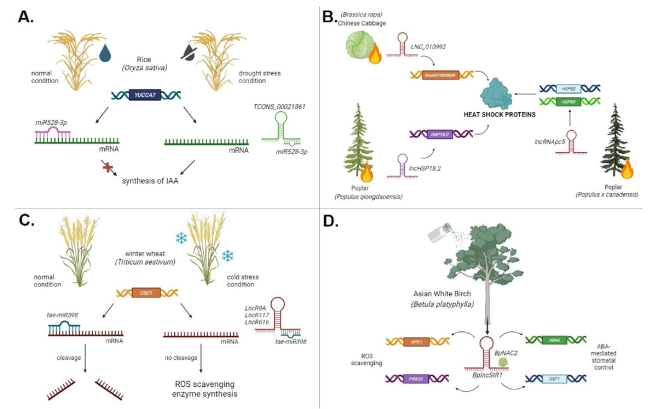
Fig. 4 Studies that investigated the role of lncRNAs in enhancing plant tolerance to abiotic stress. A The lncRNA TCONS_00021861 acts by sponging miR528-3p and preventing its binding to target mRNA, leading to increased levels of the plant growth regulator IAA to counteract the effects of drought stress in rice (Oryza sativa). B LncRNAs induced in Brassica rapa, Populus qiongdaoensis, and Populus × canadensis under heat stress regulate genes involved in the production of heat-shock proteins, which can restore misfolded proteins. C LncRNAs in wheat (Triticum aestivum) under cold stress act as ceRNAs by competing with tae-miR398 to prevent the cleavage of target mRNA, leading to the synthesis of ROS-scavenging enzymes. D The lncRNA BplncSIR1 enhances salt stress tolerance in Asian white birch (Betula platyphylla). BplncSIR1 binds to the transcription factor BpNAC2, activating genes involved in ABA-mediated stomatal control and the production of ROS-scavenging enzymes |
Heat stress
Table 2 Studies concerning specific lncRNAs that were predicted to enhance stress tolerance |
| Plant Species | Name of LncRNA | Stress | Reference | |
|---|---|---|---|---|
| 1 | Manihot esculenta (cassava) | DIR (DROUGHT-INDUCED INTERGENIC lncRNA) | drought | (Dong et al. 2022) |
| 2 | M. esculenta (cassava) | CRIR1 (cold-responsive intergenic lncRNA 1) | cold | (Li et al. 2022) |
| 3 | Gossypium hirsutum (cotton) | LncRNA973 | salt | (Zhang et al. 2019) |
| 4 | Medicago truncatula (barrelclover) | MtCIR2 | cold | (Zhao et al. 2023) |
| 5 | Pyrus spp. (pear) | HILinc1 (heat-induced long intergenic non-coding RNA 1) | heat | (Xu et al. 2020) |
Cold stress
Drought stress
Priming-induced acquired stress tolerance
Fig. 5 Melatonin enhances cadmium stress tolerance in rice (O. sativa) by lncRNA-mediated pathways. 125 lncRNAs were differentially expressed in chemically treated plants. These RNAs regulate genes that are involved in cell wall modifications to increase cadmium sequestration, maintain photosynthesis by protecting chloroplasts, and reduce oxidative damage by synthesizing antioxidants |
Table 3 Studies on the effect of chemical treatments on lncRNAs |
| Plant Species | Chemical Applied | No. of Differentially Expressed LncRNAs | Target Genes | Stress Tolerance Enhanced by Treatment | Reference |
|---|---|---|---|---|---|
| Populus × euramericana (poplar) | salicylic acid | 49 | CKX1 FBA1 | drought | (Zhang et al. 2023) |
| cold | |||||
| salt | |||||
| Fragaria × ananassa (strawberry) | ABA | 412 | - | heat | (Chen et al. 2022) |
| drought | |||||
| osmotic | |||||
| Manihot esculenta (cassava) | melatonin | 68 | - | drought | (Ding et al. 2019) |
Table 4 Studies on specific lncRNAs in Arabidopsis induced by chemical treatment |