Core
Gene & Accession Numbers
Introduction
Results
Hairy root induction and shoot regeneration
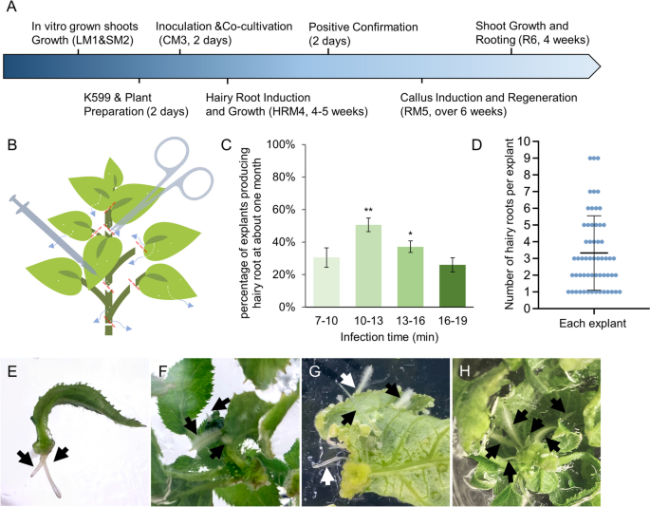
Fig. 1 The hairy root induction of kiwifruit mediated by Agrobacterium rhizogenes. A Flow diagram of Agrobacterium rhizogenes-mediated transformation (ArMT) in kiwifruit. The Blue arrow represents the process direction of transformation, and the media component and information of each step are listed in Table 1. B The pretreatment of kiwifruit explants. The white dots represent the wounds caused at the leaves, veins, and young stems, and the red lines represent the position of the cut, the blue arrows indicate where the hairy roots may occur. C Effect of different infection times on induction efficiency. Significant differences compared with each other according to one-way ANOVA followed by Tukey’s multiple comparison tests are indicated with asterisks (P < 0.001). D The number of hairy roots produced on each explant is represented by blue dots. Mean ± SD was presented. E-H The hairy roots appeared in the wound of the petiole, leaves, leaf disc, and young stems, respectively |
Table 1 Component of medium |
| Code | Medium Type | Cultivar | Additives |
|---|---|---|---|
| LM1 | In vitro grown shoots | Aca & Aeb | MSc supplemented with 0.6 mg/L NAA, 0.2 mg/L 6-BA |
| SM2 | Shoot Growth | Ac | MS supplemented with 3.0 mg/ L zeatin, 0.1 mg/ L NAA, 30 g/ L sucrose |
| Ae | MS supplemented with 2.0 mg/ L zeatin, 3.0 mg/ L 6-BA, 0.1 mg/ L IBA, 30 g/ L sucrose | ||
| CM3 | co-cultivation | Ac & Ae | 1/2 MS supplemented with 10 g/L sucrose, 100 μM Acetosyringone |
| HRM4-1 | Hairy roots induction 1 | Ac & Ae | MS supplemented with 25 g/L sucrose and 300 mg/L Cefotaximed |
| HRM4-2 | Hairy roots induction 2 | Ac & Ae | 1/2MS supplemented with 10 g/L sucrose and 300 mg/L Cefotaxime |
| RM5 | Regeneration | Ac | SM2-c supplemented with 300 mg/L Cefotaxime |
| Ae | SM2-e supplemented with 300 mg/L Cefotaxime | ||
| R6 | Rooting | 1/2 MS supplemented with 10 g/L sucrose, 0.8 mg/L IBA, 0.6% Agar |
a. Ac, Actinidia chinensis ‘Hongyang’ | |
b. Ae, Actinidia eriantha ‘White’ | |
c. MS, M519(Murashige &Skoog Basal Medium w/ Vitamins), PhytoTechnology | |
d. Cefotaxime or Timentin |
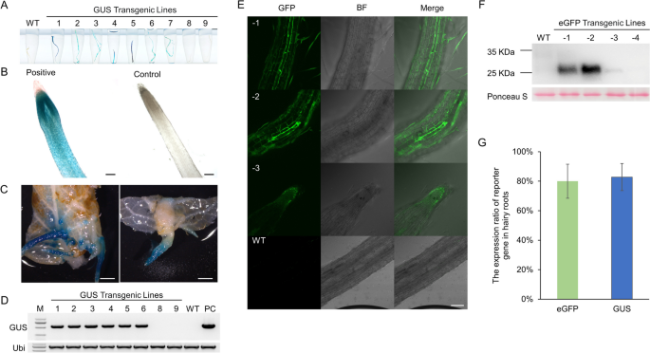
Fig. 2 Characterization of transgenic roots of GUS and eGFP. A-C The GUS staining of nine 5 weeks transgenic hairy roots (A) and enlarged field of vision under optical microscope (B) and stereomicroscope (C) from Actinidia chinensis ‘Hongyang’. B. Bar = 2 mm. C. Bars = 200 μm. D Verification of T-DNA insertion fragment of GUS by PCR amplification. M, marker. WT, wild type. PC, positive control. E The eGFP fluorescence signal in 5 weeks transgenic hairy roots of Actinidia eriantha ‘White’. c, root tip. d, the wild type. Bars = 200 μm. F Western blotting with anti-GFP antibodies of 4 transgenic lines and wild type. Molecular mass markers are shown on the left. The expected size of eGFP is 26.9 KDa. G The expression ratio of eGFP and GUS gene in hairy roots. Data shown are averages ±SD; n > 15 |
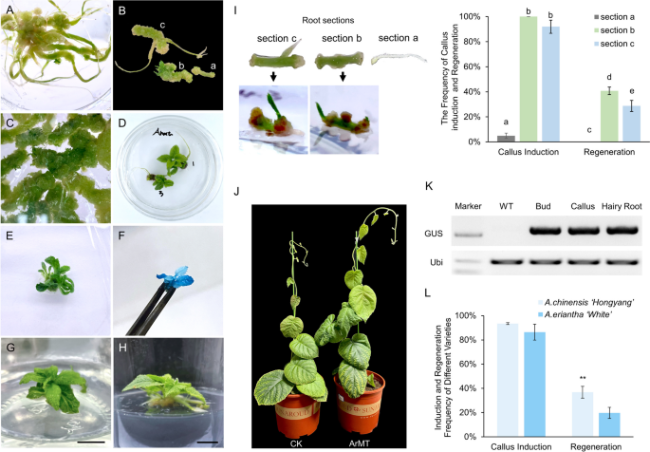
Fig. 3 Callus induction and regeneration of hairy roots through a removing-root-tip method. A Callus induced in the presence and (B) removal of root tip; a, segment near the tip removing the root tip; b, mature zone; c, the zone with lateral roots. C Regenerative buds are produced during early induction on sections without the root tip. D The root segment of the root tip is removed to induce callus and regeneration. E F K GUS transgenic hairy root regeneration seedling and chemical staining, and GUS gene T-DNA insertion verification. G H Growth and rooting of regenerated seedlings induced by hairy roots. Bar = 1 cm. I The different sections of hairy root (section a, root tip; section b, the elongation zone; section c, the maturation zone with lateral roots) showed the different frequencies of callus induction and regeneration. J The WT and transgenic plants of 35S::GUS were grown in soil for 2 months. L Statistics of callus induction frequency and regeneration frequency from different varieties of kiwifruit hairy roots. Data shown are averages ±SD; n > 50 and asterisks indicated significant differences at P < 0.001 |
A.Rhizogenes-mediated gene editing of AcCEN4 and AeCBL3
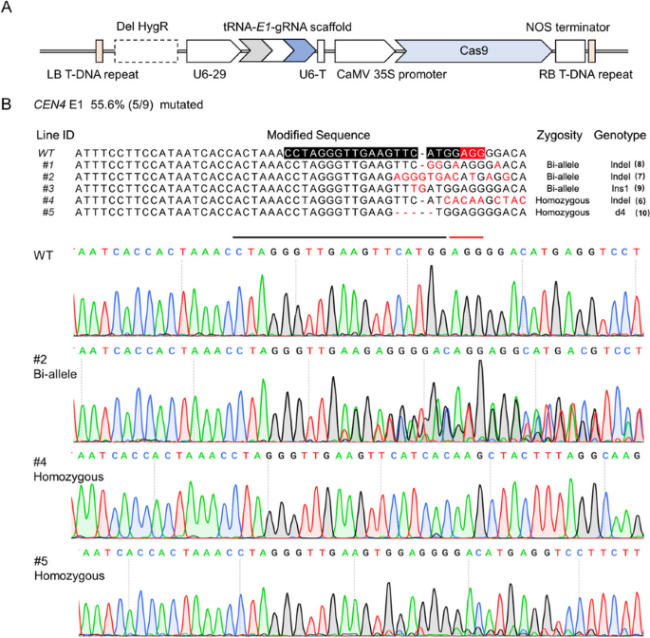
Fig. 4 Agrobacterium rhizogenes-mediated gene editing of CEN4 gene in kiwifruit. A Polycistronic tRNA-gRNA (PTG)/Cas9 vector structure targeting the CEN4-E1 site, the screening marker gene was removed and represented by a black dotted box, the gray arrow, white arrow and blue arrow in the PTG structure represent tRNA, sgRNA and gRNA scaffold, respectively. B Edited results of CEN4 gene target region sequences in 9 transgenic hairy roots, of which 5 were detected editing. Black and red indicate the target sequence and PAM region. Indel, small fragment insertion and deletion variation; Ins, insertion; d, deletion; WT, wild type. The number in brackets indicates the clones of PCR products for Sanger sequencing |
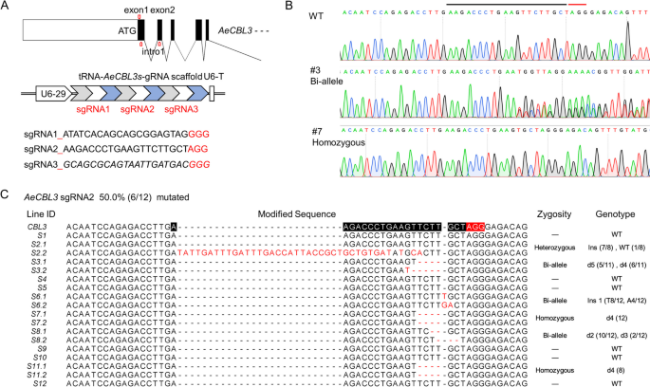
Fig. 5 The gRNA screening and gene editing of AeCBL3. A Design of 3 targeted sequences on the first exon and the second exon of the AeCBL3 gene represented by red arrows. The gray arrow, white arrow, and blue arrow in the PTG structure represent tRNA, sgRNA, and gRNA scaffold, respectively. The multiple editing structure is composed of three tRNA-gRNA combinations. B Genome sequence of the sgRNA2 site region in the transgenic hairy root, the black and red lines represent the targeted sequence and the PAM region. C Gene modification of 12 independent transformed hairy roots, and genotype classification. Monoclonal sequencing of each independently transformed hairy root was performed to analyze specific gene editing forms, all the modified sequences were displayed, while the genome sequences of other lines were not changed. Target sequences and PAM regions were labeled with black and red lines. Ins, insertion; d, deletion; WT, wild type. The number in brackets (a/b), a represents the number of colonies of this type, b represents the total number of colonies |
AeCBL3 mediates the formation of calcium oxalate crystals
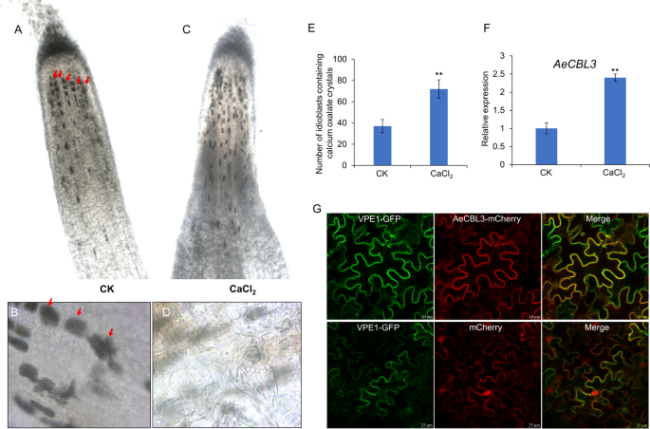
Fig. 6 CaCl2 treatment increased the formation of calcium oxalate crystals in the root tip. A-D Microscopy observation of primary root of ‘White’ treated without (A) and with 15 mM CaCl2 (C) for 1 week. Red arrows indicated the crystal idioblasts. The bundled needle-shaped raphide crystals in the idioblasts (B) and the released raphide crystals from broken idioblasts (D). E The counted number of crystal idioblasts in root tips of ‘White’ treated without (CK) and with CaCl2 Data shown are averages ±SD; n > 10. ‘**’ indicates significant differences at P < 0.001. F qRT-PCR analysis of AeCBL3 in the roots treated without (CK) or with CaCl2. The three independent experiments were performed and three biological replicates for each treatment. ‘**’ indicates significant differences at P < 0.001. G Confocal microscope observation of AeCBL3-mCherry, mCherry, and vacuolar membrane marker protein VPE1-GFP transiently expressed in the tobacco leaves |
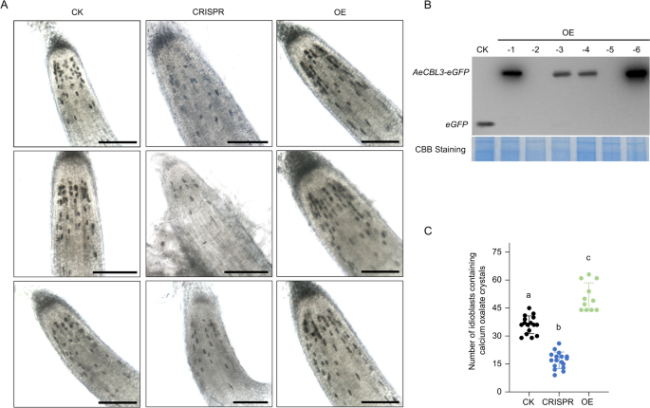
Fig. 7 AeCBL3 mediates the formation of calcium oxalate crystal idioblasts in kiwifruit. A Observation of idioblasts in hairy roots under an optical microscope. CK, Control lines; CRISPR, AeCBL3-edited lines; OE, Overexpression lines. Bar = 200 μm. B Western blot analysis of CK and OE lines using anti-GFP antibodies. C The number of idioblasts containing calcium oxalate crystals in the independent lines of Control, Overexpression, and CRISPR was counted, which was represented by black dots, green dots, and blue dots, respectively. Data shown are averages ±SD; n > 10. Significant differences compared with each other according to two-way ANOVA followed by Tukey’s multiple comparison tests are indicated with a, b, and c (P < 0.001) |