Core
Gene and accession numbers
Introduction
Result
Identification of PbrADC1 as the candidate gene involved in the development of superficial scald in pear
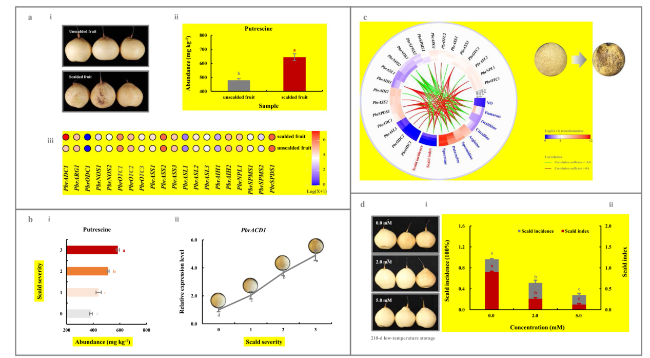
Fig. 1 Characterization of PbrADC1 gene involved in superficial scald development via regulating putrescine biosynthesis. a Alternation of putrescine metabolism upon superficial scald development in ‘Yali’ fruit. (a-i) Visual quality change. (a-ii) Putrescine level. (a-iii) Expression profiles of 21 putrescine-metabolism-related genes. ‘Yali’ fruit, with and without superficial scald, were sampled after -0.5 ℃ for 180 d followed by a 7-d shelf life at 20 ℃. b Putrescine content and PbrADC1 expression abundance in ‘Yali’ fruit of different superficial scald severities. (b-i) Putrescine content. (b-ii) PbrADC1 mRNA abundance. ‘Yali’ fruit, with different superficial scald severities (Hui et al. 2016), were sampled after -0.5 ℃ for 180 d followed by a 7-d shelf life at 20 ℃. The expression abundance of PbrADC1 in the unscalded fruit (Severity 0) was set as 1.0 for RT-qPCR assay. c Dynamic change of putrescine metabolism during cold storage of ‘Dangshansuli’ fruit. ‘Dangshansuli’ fruit were sampled every 60-d storage at 0.5 ℃ followed by a 7-d shelf life at 25 ℃. d Impact of exogenous putrescine treatment on superficial scald development in pear fruit. (d-i) Visual quality change. (d-ii) Superficial scald incidence and index. ‘Yali’ fruit were randomly divided into three groups for different treatments: 0.0 (H2O), 2.0-mM, and 5.0-mM putrescine immersion for 15 min before 210-d storage at -0.5 ℃ followed by a 7-d shelf life at 20 ℃. A total of 21 putrescine-metabolism-related genes were characterized from the P. bretschneideri Rehd. genome (Table S2). Data represent the mean values of three biological replicates, except for the transcriptome assay of the (un) scalded ‘Yali’ fruit (two replicate). Different lowercase letters meant significance between samples (p < 0.05). Color scale represented normalized log2-transformed (FPKM + 1), where red indicated a high level, blue represented a low level, and white indicated a medium level. Absolute correlation coefficients between attributes ≥ 0.8 were visualized in the heatmap, where red lines demonstrated extremely strong positive correlations, while green demonstrated extremely strong negative associations |
Functional validation of PbrADC1 involved in putrescine biosynthesis and thus chilling resistance
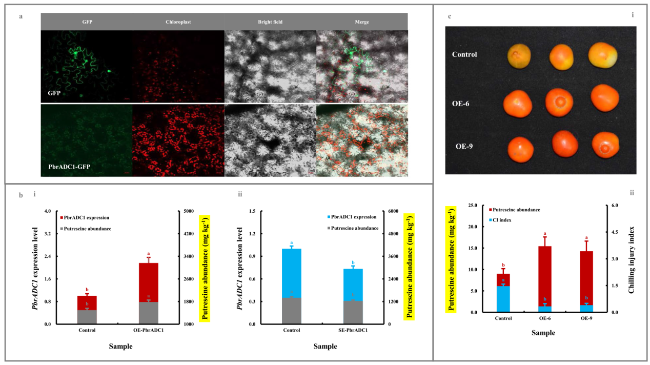
Fig. 2 Functional validation of PbrADC1 involved in fruit putrescine biosynthesis and thus chilling resistance. a Subcellular localization of PbrADC1. The recombinant pBI221-PbrADC1 vector was transformed into N. benthamiana leaves before fluorescence signal detection. b Impact of transient genetic transformation in the ripe ‘Dangshansuli’ fruit on putrescine abundance. (b-i) Transient overexpression of PbrADC1 gene. The ripe ‘Dangshansuli’ fruit transformed with the empty vector was used as a control. (b-ii) Transient silence of PbrADC1 gene. The ripe ‘Dangshansuli’ fruit co-transformed with empty pTRV2 and pTRV1 was used as a control. The expression abundance of PbrADC1 in control fruit was set as 1.0 for RT-qPCR assay. c Impact of overexpressing PbrADC1 gene in tomato on fruit putrescine biosynthesis and chilling resistance. (c-i) Visual quality change. (c-ii) Putrescine level and chilling injury index. Tomato fruits at 35 DAFB, including the wide-type (control) and the PbrADC1-overexpressing (OE) lines, were harvested and then exposed to 4 ℃ for 10 d followed by 20 ℃ storage for 7 d. Data represented the mean value of three biological replicates, and different lowercase letters meant significance between samples (p < 0.05) |
Evolution and characteristics of plant ADCs
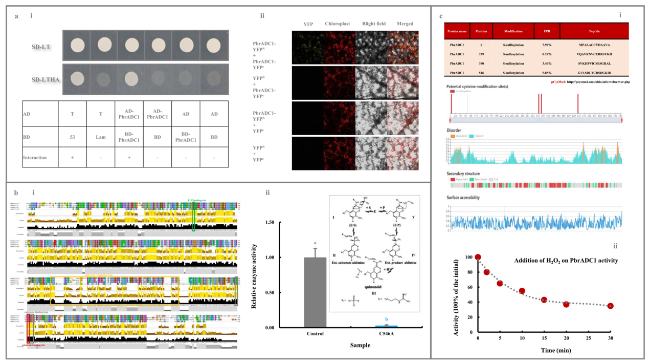
Fig. 3 Characteristics of PbrADC1. a PbrADC1 self-interaction determination. (a-i) Y2H assay. Transformants containing AD-T & BD-53, AD-T & BD-Lam, AD & BD, AD & BD-PbrADC1, and AD-PbrADC1 & BD were used as controls. (a-ii) BiFC assay. Transformants containing YFPN & YFPC, YFPN & PbrADC1-YFPC, and YFPC & PbrADC1-YFPN were used as controls. b Identification and functional validation of the substrate-binding residue Cys546 in PbrADC1. (b-i) Alignment of PbrADC1 with AtADCs and HpADC1 by Jalview Version 2. Information on AtADCs and HpADC1 was summarized by Hanfrey et al. (2001). (b-ii) Functional validation of Cys546. Enzyme activities of the His-tagged recombinant PbrADC1 and PbrADC1.C546A proteins were analyzed based on the method of Song et al. (2010). c Exogenous H2O2 treatment on PbrADC1 activity. (c-i) Identification of the H2O2-modified Cys residues in PbrADC1. The H2O2-modified Cys residues in PbrADC1 was predicted by pCysMod database (Li et al. 2021b). (c-ii) Impact of exogenous H2O2 treatment on PbrADC1 activity. The residual activity was expressed as a percentage of the initial (0 min). Data represented the mean value of three biological replicates, and different lowercase letters meant significance between samples (p < 0.05) |
Characterization and confirmation of PbrWRKY62 as the upstream regulator of PbrADC1 gene
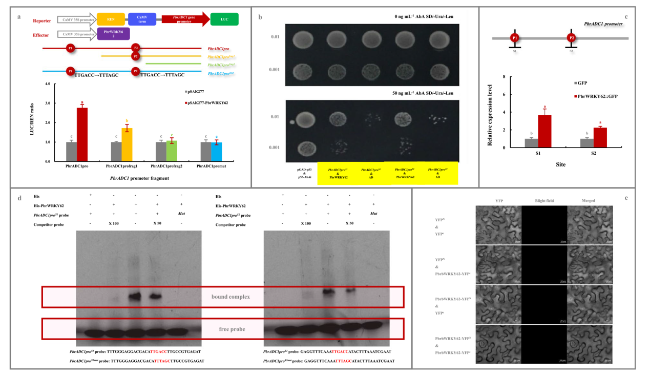
Fig. 4 Confirmation of PbrWRKY62 as the upstream regulator of PbrADC1 gene. a Dual-luciferase assay. Transformants containing empty pSAK277 and each report vector were used as controls. b Y1H assay. Yeast cell co-transformed with pGAD7-p53 & p53-AbAi was used as a positive control, while yeast cell with pGADT7-AD & PbrADC1proS1-pAbAi and pGADT7-AD & PbrADC1proS2-pAbAi, respectively, were used as negative controls. c ChIP-qPCR analysis. Pear calli overexpressing the empty pCAMBIA1300-GFP plasmid was used as a negative control. d In vitro EMSA assay. FAM luciferase-labelled PbrADC1 promoter fragments, containing the W-box elements (TTGACC) and its mutant (TTGACC → TTTAGC), were named as PbrADC1proS1 probe, PbrADC1proS2 probe, PbrADC1proS1mut probe, and PbrADC1proS2mut probe, respectively, and the unlabeled PbrADC1 promoter fragments containing the W-box elements were used as competitor probes. The presence and absence of His protein, His-PbrWRKY62 protein, labeled probe, or competitor probe were indicated by “ + ” and “ − ”, respectively. Competitor probe concentrations were 50-fold (50 ×) and 100-fold (100 ×) those of the labeled probe. e BiFC assay for the self-interaction of PbrWRKY62. Transformants containing YFPN & YFPC, YFPN & PbrWRKY62-YFPC, and YFPC & PbrWRKY62-YFP.N were used as controls. Data represented the mean value of three biological replicates, and different lowercase letters meant significance between samples (p < 0.05) |
Functional validation of PbrWRKY62 involved in putrescine biosynthesis and thus chilling resistance
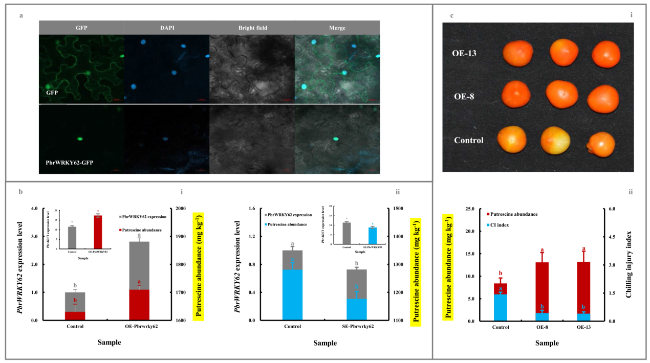
Fig. 5 Functional validation of PbrWRKY62 involved in fruit putrescine biosynthesis and thus chilling resistance. a Subcellular localization of PbrWRKY62. 4’,6-diamidino-2-phenylindole (DAPI) was used as a nuclear indicator (Kapuscinski 1995). b Impact of transient genetic transformation in the ripe ‘Dangshansuli’ fruit on putrescine abundance. (b-i) Transient overexpression of PbrWRKY62 gene. The ripe ‘Dangshansuli’ fruit transformed with the empty vector was used as a control. (b-ii) Transient silence of PbrWRKY62 gene. The ripe ‘Dangshansuli’ fruit co-transformed with empty pTRV2 and pTRV1 was used as a control. The expression level of PbrWRKY62 in control fruit was set as 1.0 for RT-qPCR. c Impact of overexpressing PbrWRKY62 gene in tomato on fruit putrescine biosynthesis and thus chilling resistance. (c-i) Visual quality change. (c-ii) Putrescine content and chilling injury index. Tomato fruits at 35 DAFB, including the wide-type (control) and overexpressing (OE) lines, were harvested and then exposed to 4 ℃ for 10 d followed by 20 ℃ storage for 7 d. Data represented the mean value of three biological replicates, and different lowercase letters meant significance between samples (p < 0.05) |
Discussion
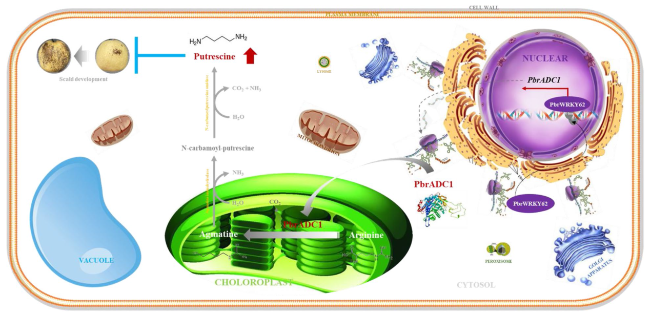
Fig. 6 Schematic model of PbrWRKY62 and its downstream target gene PbrADC1 elevating fruit putrescine level and thus chilling tolerance. PbrWRKY62, located in the nucleus, could interact with two W-box elements in PbrADC1 promoter as monomer and then activate its expression. After translation in the ribosome, PbrADC1 was then transported into the chloroplast, where it converted arginine to agmatine, elevating putrescine level and thus chilling resistance of fruit |