Core
Gene and accession numbers
Introduction
Results
Seed development in the large-seeded cultivar 'HZ' and the abortive-seeded cultivar 'NMC'
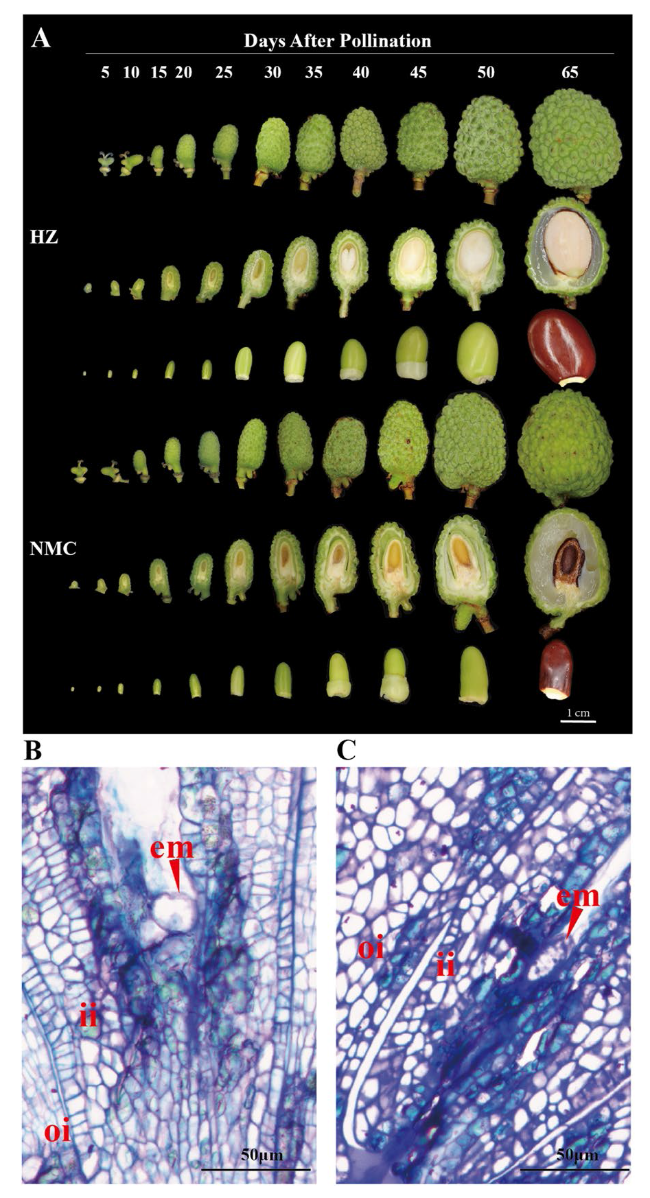
Fig. 1 Comparison of seed development between the large-seeded cultivar 'HZ' and the abortive-seeded cultivar 'NMC’. A Photos showing the different stages of seed development after pollination (DAP) in ‘HZ’ and ‘NMC’. Bar = 1 cm; The cross-sections of seed at 5 DAP in ‘HZ’ (B) and ‘NMC’ (C). The red arrows indicate the embryos. em, embryo; ii, inner integument; oi, outer integument. Scale bars = 50 μm |
DNA methylome characteristics of 'HZ' and 'NMC' seeds at an early developmental stage
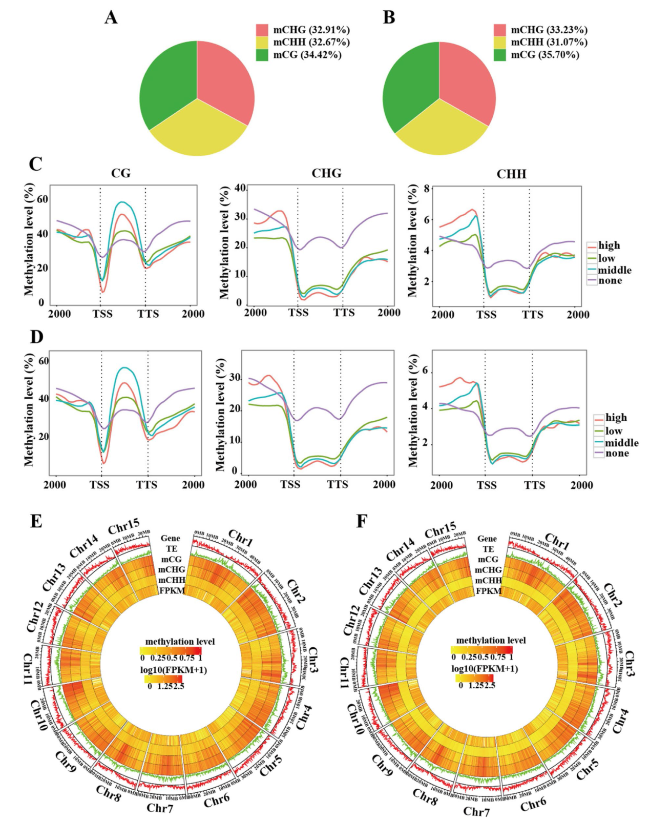
Fig. 2 Genome-wide DNA methylation profiling during the early stages of seed development in ‘HZ’ and ‘NMC’. A pie chart indicating the the proportion of mCG, mCHG and mCHH in all methylcytosines (mCs) at 5 DAP in 'HZ' (A) and 'NMC' (B). The DNA methylation levels within the gene body and ± 2 kb regions of genes are shown for different sequence contexts in gene sets that are expressed at different levels at 5 DAP in 'HZ' (C) and 'NMC' (D). The expression levels are categorized as high (FPKM > 100), middle (10 < FPKM ≤ 100), low (1 < FPKM ≤ 10), and none (FPKM ≤ 1). The terms TSS and TTS represent the transcription starting site and transcription terminal site, respectively. A chromosome heat map illustrating gene density, transposable elements density, DNA methylation levels in different sequence contexts, and gene expression levels of seeds at 5 DAP in 'HZ' (E) and 'NMC' (F). The bin size used for this visualization is 100 kb. The outer ring displays the chromosome names and scale |
DNA methylation levels in ‘HZ’ were higher than ‘NMC’ seeds at an early developmental stage
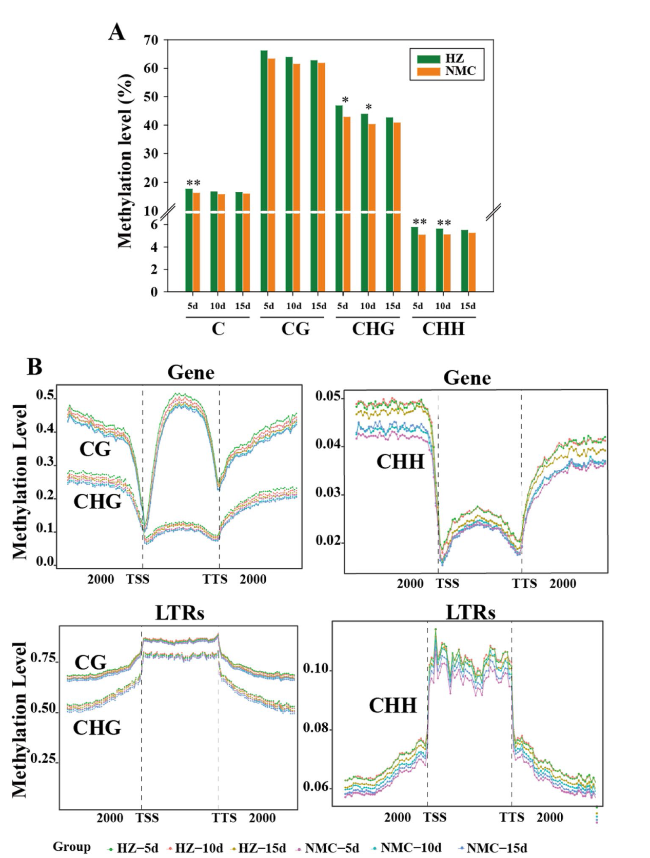
Fig. 3 DNA methylation level in ‘HZ’ seeds was higher than those in ‘NMC’ seeds during the early stages of seed development. A DNA methylation levels (%) of CG, CHG, and CHH sequence contexts are represented respectively as vertical bar charts. Asterisks indicate a significant difference (t-test: * P < 0.05, ** P < 0.01). B DNA methylation levels in the gene body, upstream and downstream (2 kb) regions of genes, and long terminal repeats (LTRs) transposable elements in different sequence contexts (CG, CHG, and CHH) at 5 DAP, 10 DAP and 15 DAP in ‘HZ’ and ‘NMC’ |
Differentially methylated regions and differentially expressed genes are enriched in the ROS pathway
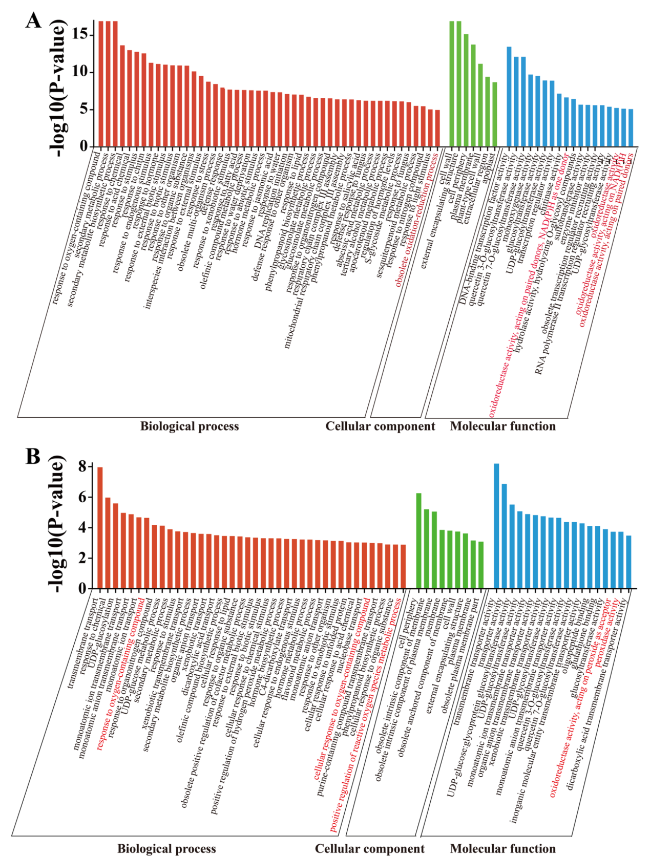
Fig. 4 GO term enrichment analysis of genes in DMRs or DEGs. A GO term enrichment analysis of the differentially expressed genes (DEGs). B GO term enrichment analysis of genes in differentially methylated regions (DMRs) in CHH. Words with the red color indicate the biological process associated with the oxidation reaction pathway |
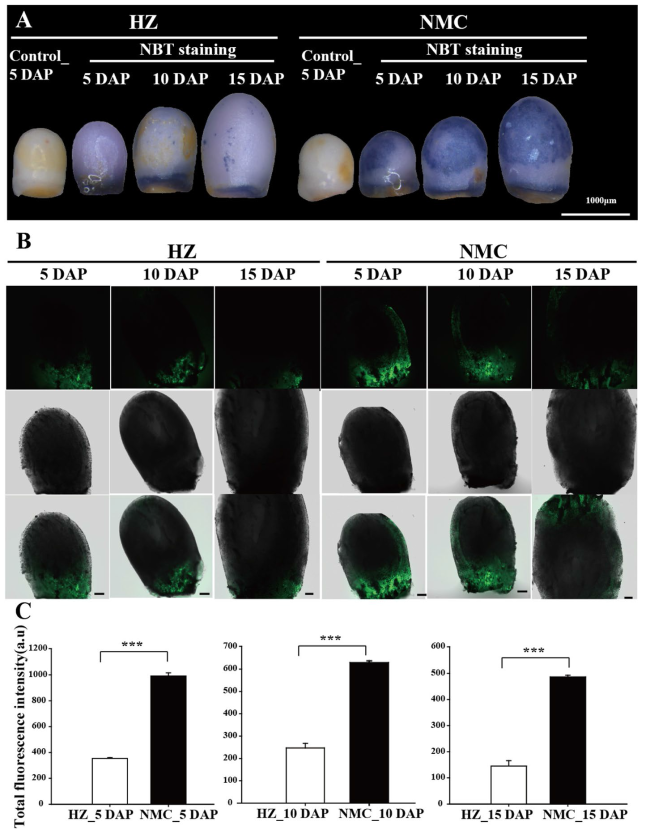
Fig. 5 Comparable assays of reactive oxygen species production in the seeds between ‘HZ’ and ‘NMC’. A Nitroblue tetrazolium (NBT) was used to detect the accumulation of superoxide and hydrogen peroxide (H2O2) in the ‘HZ’ and ‘NMC’ seeds at 5 DAP, 10 DAP, and 15 DAP. Scale bars are 1000 μm; B Dichlorodihydrofluorescein (DCF) fluorescence of ROS in longitudinal sections of the seeds between 'HZ' and 'NMC'. Scale bars are 100 μm. C Quantification of the DCF fluorescence intensity in the seeds of 'HZ' and 'NMC’. At least three independent experiments with three samples each were performed. Asterisks indicate a significant difference (Independent-Sample t-test: *** P < 0.001) |
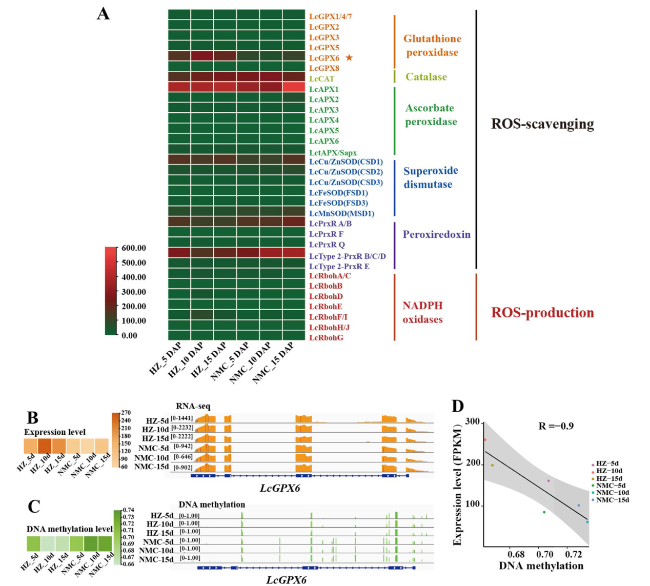
Fig. 6 Expression patterns of LcGPX6 in ‘HZ’ and ‘NMC’ seeds. A A heat map showing the expression patterns of genes involved in the production and scavenging of ROS in the seeds of ‘HZ’ and ‘NMC’. B A heat map (left panel) and genome browser view (wiggle plots) (right panel) indicate that the expression level of LcGPX6 was higher in ‘HZ’ seeds than that in ‘NMC’ seeds at 5 DAP, 10 DAP, and 15 DAP. C A heat map (left panel) and genome browser view (vertical lines) (right panel) indicate that the DNA methylation level of LcGPX6 was lower in ‘HZ’ seeds than that in ‘NMC’ seeds at 5 DAP, 10 DAP, and 15 DAP. D The correlation analysis of expression level and DNA methylation level of LcGPX6 |
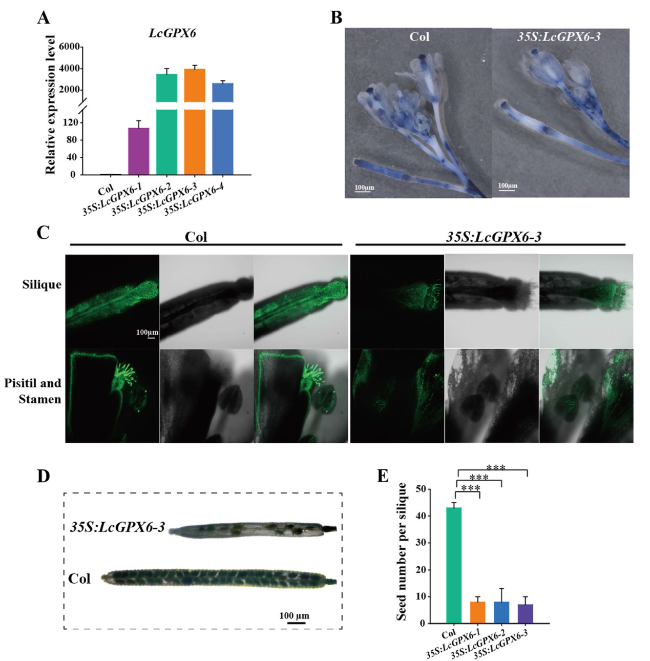
Ectopic expression of LcGPX6 in Arabidopsis causes defects in seed development
Fig. 7 Ectopic expression of LcGPX6 in Arabidopsis causes defects in seed development. A Ectopic expression levels of LcGPX6 in wild type Col and different transgenic lines. qRT-PCR was performed and the Ubiquitin 10 from Arabidopsis was used as the internal reference to analyze the relative level of gene expression. B NBT was used to detect the superoxide and H2O2 accumulation in the inflorescences of Col and 35S:LcGPX6-3 transgenic plants. Scale bars are 1000 μm. C DCF fluorescence images of silique, pisitil and stamen between the Col and 35S:LcGPX6-3 transgenic plants. Scale bars are 100 μm. D Phenotype of the siliques and seeds in wild type Col and 35S:LcGPX6-3 transgenic plants. Scale bars are 100 μm. E Quantitative assay of seed number per silique in wild type Col and LcGPX6 transgenic lines. Error bar indicates mean ± SD (n = 10 siliques). Statistical significance was determined via employing Independent-Sample t-test (***P < 0.001) |
Discussion
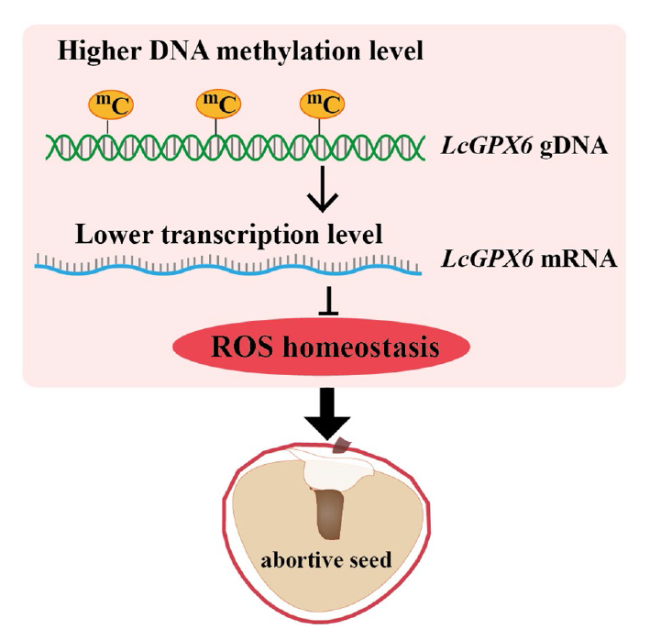
Fig. 8 Proposed model of DNA methylation-mediated ROS production contributes to seed abortion in litchi. During the seed development of seed-abortive cultivars in litchi like 'NMC', it was observed that the genomic DNA of LcGPX6 exhibited relatively higher levels of DNA methylation. This higher methylation resulted in a lower expression level of LcGPX6. Since LcGPX6 is a key gene that encodes an enzyme responsible for scavenging reactive oxygen species (ROS), the decreased expression level of LcGPX6 may lead to an imbalance in ROS homeostasis, therefor resulting in the production of abortive seeds within the fruit |