Core
Gene and Accession Numbers
Introduction
Results
Assembly and annotation of the haplotype genome of R. chinensis ‘Chilong Hanzhu’
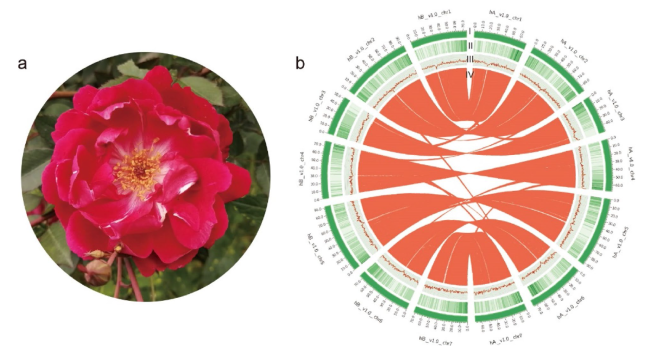
Fig. 1 The features of Rosa chinensis ‘CH’ genome. a The plant of R. chinensis ‘CH’. b Distribution of ‘CH’ genomic features. (I) Circular representation of the 14 chromosomes of two haplotype genomes, length in Mb, (II) gene density, (III) GC content in 500 kb windows, and (IV) each linking line in the center of the circle connects a pair of homologous genes between the two haplotype genomes of ‘CH’ |
Table 1 Statistics for R. chinensis ‘CH’ Haplotype genomes |
| HapA | HapB | |
|---|---|---|
| Contig N50 | 10.89 Mb | 18.05 Mb |
| scaffolds N50 | 69.83 Mb | 74.66 Mb |
| Longest contig | 92,402,791 bp | 92,644,500 bp |
| Chromosome number | 7 | 7 |
| Number of contigs | 342 | 264 |
| Number of scaffolds | 193 | 176 |
| Genome-size | 518,698,571 bp | 541,343,551 bp |
| Rate of anchoring (%) | 97.9 | 98.4 |
| GC content (%) | 38.95 | 38.84 |
| BUSCO_genome (%) | 98.4 | 98.4 |
| TE (%) | 59.6 | 61.83 |
| Number of predicted genes | 31,744 | 32,340 |
| BUSCO_protein (%) | 98.6 | 99.0 |
Genetic variation of the two haplotype genomes in highly heterozygous ‘CH’
Fig. 2 Genetic variation of the haplotype genome in high heterozygous R. chinensis ‘CH’. a-c The length distribution of duplications (a), inversions (b) and translocations (c) between the two haplotypes genomes. d-e The genomic structural variation characteristics between the two haplotype genomes. hA and hB mean the haplotype A and haplotype B of ‘CH’ genome, separately. f. Collinearity and structural variation distribution between two haplotypes of ‘CH’. g. Differential expression ratios of alleles in four tissues of roots, stems, leaves, and flowers |
Phylogenetic evolution
Fig. 3 Evolution of the R. chinensis ‘CH’ and influence of family gene expansion on the main components of floral aroma. a Phylogenetic tree for ‘CH’ hB and ten other eudicot species contained three outgroups. Gene family expansions are indicated in green, and contractions in black; The estimated divergence time (million years ago, MYA) is indicated at each node; numbers in brackets are the 95% confidence intervals (each center is defined as mean value). The red dot represents a calibration point. b-c Gene Ontology (GO) enrichment analysis of expanded gene families in ‘CH’ (b) and ‘OB’ (c). d Accumulation histogram of four groups of volatile organic compounds of R. chinensis ‘OB’ and ‘CH’ |
Identification of pigments and differentially expressed genes in red and pink petals
Fig. 4 Potential anthocyanin regulatory pathway of red or pink petals in R. chinensis. a The metabolites were identified by LC_MS in ‘CH’ and ‘OB’ petals. Peak 1-3 represent cyanidin-3,5-diglucoside, pelargonidin-3,5-O-diglucoside and cyanidin-3-glucoside, respectively. b Proposed biosynthetic pathway of anthocyanin synthesis in rose. c The different 35S:RcMYB114b transgenic lines of rose turned red. d Expression of RcMYB114b (left) and anthocyanin biosynthesis pathway genes (right) in different 35S:RcMYB114b transgenic lines of rose callus. The bars indicate the SD. Stars mean significant differences (*** represents P < 0.001, ** represents P < 0.01, * represents P < 0.05) analyzed by one-way ANOVA. e. LC-MS analysis of 35S:RcMYB114b transgenic rose callus and control. Peak 1-3 represent cyanidin-3,5-diglucoside, pelargonidin-3,5-O-diglucoside and cyanidin-3-glucoside, respectively. f. The transient expression of RcMYB114b in apple fruit. The black triangle represents the site of the injection. g. Expression of anthocyanin biosynthesis pathway genes in RcMYB114b transgenic lines of apple. The bars indicate the SD. Stars mean significant differences (*** represents P < 0.001, ** represents P < 0.01, * represents P < 0.05) analyzed by one-way ANOVA |
RcMYB114b contributes to the formation of petal colour
Multi-TFs affect the response of rose petal number and size to high temperature
Fig. 5 Changes in the number and size of petals in R. chinensis ‘CH’ under high temperature treatment. a Flower morphology at different stages under high temperature (downer) and normal temperature (upper) treatment. b Statistics of the number and size of floral organs under different temperature treatments. Stars (***) mean significant differences (P < 0.001) analyzed by two-way ANOVA. c The cell density middle of petals under high temperature (right) and normal temperature (left) treatment. d-e The expression of RcANT1, RcDA1, RcAG1, RcAG2 and RcSVP1. Stars mean significant differences (*** represents P < 0.001, ** represents P < 0.01, * represents P < 0.05) analyzed by T-test. f. The coverage of RcAP2Lwt exon 10th with miR172 binding site under high temperature treatment. Stars mean significant differences (* represents P < 0.05) analyzed by T-test. g. The dual-LUC assays. Red represents higher signal intensity and blue represents lower signal intensity. The bars indicate the SD. Stars (***) mean significant differences (P < 0.001) analyzed by one-way ANOVA |