Core
Gene & accession numbers
Introduction
Results
SA promotes leaf senescence and ROS burst in cucumber
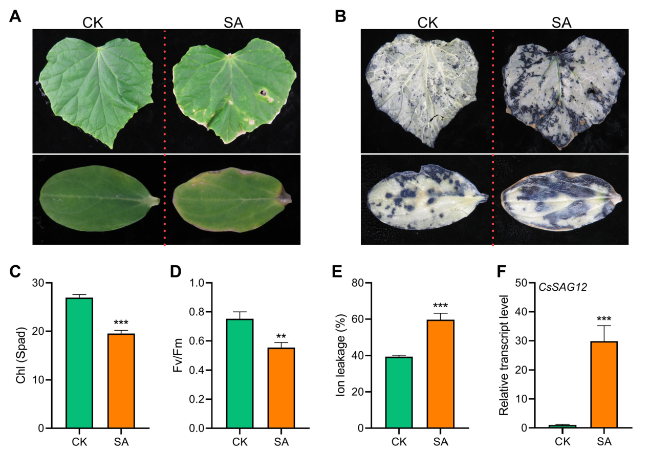
Fig. 1 SA promotes leaf senescence and ROS burst in cucumber. A Phenotypes of SA and H2O (CK)-treated cucumber leaves and cotyledons. B Phenotypes of NBT-stained cucumber leaves and cotyledons after SA and CK treatments. C-F Chlorophyll content (C), Fv/Fm ratio (D), ion leakage (E), and relative transcript level of CsSAG12 (F) in SA- and CK-treated cucumber leaves. In (A-F), detached leaves and cotyledons were floated on 1mM SA solution for 5 days, data are means ± SD (n = 3 biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001 (t-test), primers are listed in Table S1 |
SA regulates chlorophyll degradation and ROS production by up-regulating CsCCGs and CsRBOHB
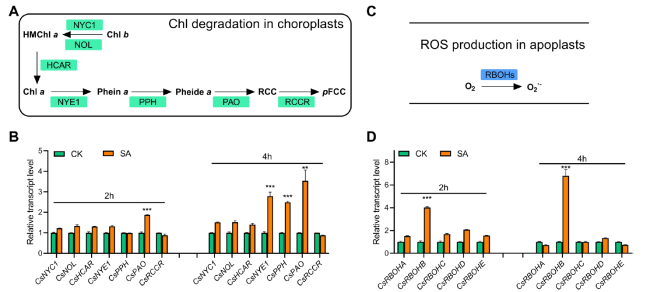
Fig. 2 SA up-regulates the expression of CsCCGs and CsRBOHB. A The biochemical pathway of chlorophyll degradation in chloroplasts. B Time-course expression patterns of CsCCGs in response to SA treatment. C The biochemical pathway of ROS biosynthesis in apoplasts. D Time-course expression patterns of CsRBOHs in response to SA treatment. Data are means ± SD (n = 3 biological replicates), **P < 0.01, ***P < 0.001 (t-test), primers are listed in Table S1 |
SA signaling primarily activates WRKY transcription factors in cucumber
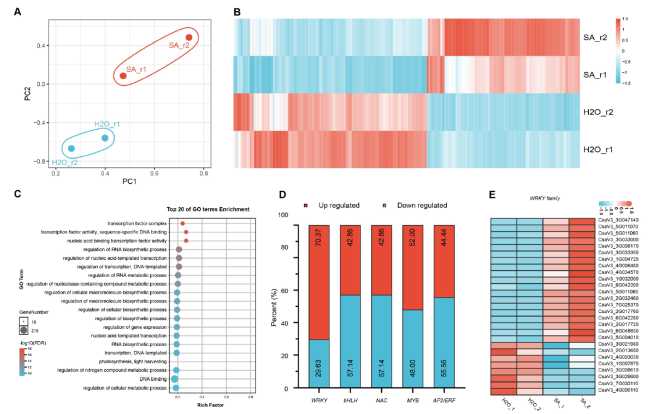
Fig. 3 SA-activated signal transduction involves WRKY transcription factors. A Principal component analysis (PCA) of SA- and H2O-treated cucumber leaves. B The heatmap of DEGs between SA- and H2O-treated samples (n = 2 biological replicates). C GO analysis of SA-regulated DEGs. D A 100% stacked bar chart, showing the SA-regulated transcription factors in cucumber. E The heatmap of SA-regulated WRKY family genes in cucumber leaves. DEGs were defined on the basis of cut values Log2 (foldchange) ≥ 1, FDR < 0.05, samples were treated with SA and H2O for 30 min (n = 2 biological replicates) |
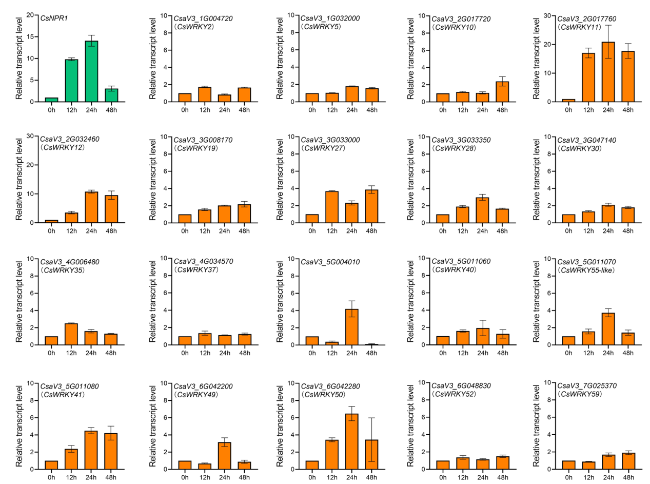
CsNPR1 robustly induces the expression of CsWRKY11
Fig. 4 CsNPR1 regulates the expression of CsWRKYs. Time-course expression patterns of CsWRKYs after CsNPR1 being over-expressed in cucumber cotyledons. Relative transcript levels were calculated as the ratio of those measured in p35S::CsNPR1- over those in empty vector-transformed cotyledons, n = 3 biological replicates |
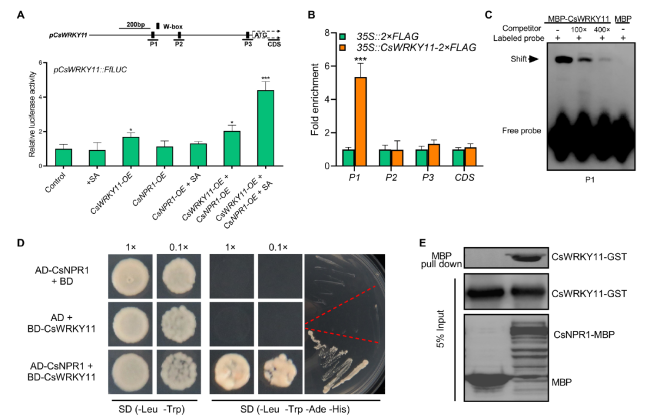
CsWRKY11 and CsNPR1 directly interact on the promoter of CsWRKY11 to activate its transcription
Fig. 5 CsNPR1 promotes the transcriptional activation of CsWRKY11 on its promoter. A Dual-luciferase analysis of the effects of CsWRKY11 and CsNPR1 on the promoter activity of CsWRKY11 (pCsWRKY11::FfLUC). Arabidopsis protoplasts were co-transformed with pCsWRKY11::FfLUC and p35S::CsWRKY11, p35S::CsNPR1 or empty vector (control) alone or in combination, LUC activity was monitored 16 h post culturing. B ChIP-qPCR assay of the binding of CsWRKY11 to its encoding gene promoter. p35S::CsWRKY11-2 × FLAG- and p35S::2 × FLAG-transfected cucumber cotyledons were used for the analysis. Enrichment of the target fragments was normalized to CsACTIN, the coding region was used as an internal control. C EMSA of the interaction between CsWRKY11 and W-box on its encoding gene promoter. A CsWRKY11 promoter fragment containing a W-box (P1) was biotin-labeled as a probe, with the same fragment unlabeled being used as a competitor. D Physical interaction of CsNPR1 and CsWRKY11 in the Y2H assay. The pGADT7-CsNPR1 and pGBKT7-CsWRKY11 plasmids were co- transferred into yeast strain AH109, and positive clones was grown and screened on a quadruple dropout (QDO: -Leu, -Trp, -Ade, -His) medium. pGADT7-CsNPR1 + pGBKT7 and pGADT7 + pGBKT7-CsWRKY11 transformed yeast cells were used as negative controls. E Physical interaction of MBP-CsNPR1 and GST-CsWRKY11 in the pull-down assay. Coding sequences of CsNPR1 and CsWRKY11 were inserted into pMAL-c5g and pGEX4T-1 plasmids respectively; plasmid-transformed Escherichia coli (BL21) cells were used for protein preparation. Input and pulled-down proteins were detected with anti-GST and anti-MBP antibodies |
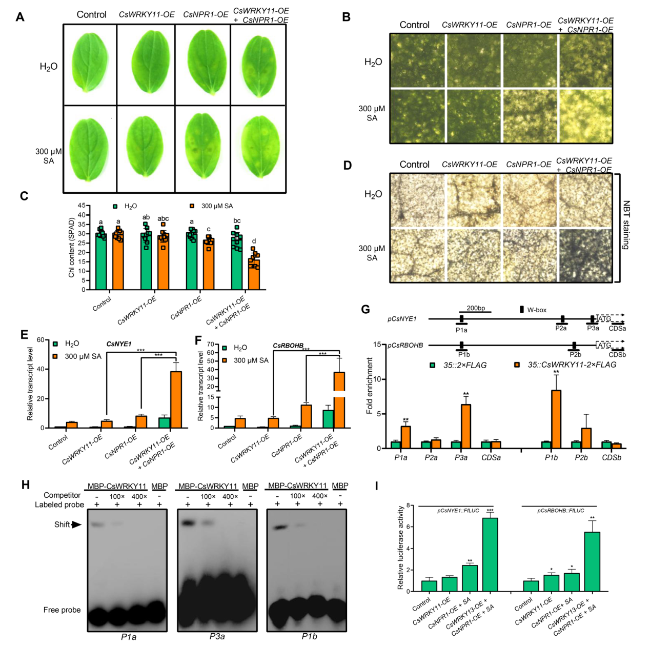
CsNPR1 and CsWRKY11 synergistically promote SA-induced chlorophyll degradation and ROS burst
Fig. 6 CsWRKY11 and CsNPR1 synergistically promote chlorophyll degradation and ROS biosynthesis. A De-greening phenotypes of cucumber cotyledons after CsWRKY11 and/or CsNPR1 being transiently over-expressed, in the presence or absence of SA. B Microscopic images of the leaf tissues around Agrobacterium tumefaciens-infiltrated sites on CsWRKY11- and/or CsNPR1-overexpressed cotyledons, in the presence or absence of SA. C Chlorophyll contents in CsWRKY11- and/or CsNPR1-overexpressed cucumber cotyledons, in the presence or absence of SA. D Microscopic images of the leaf tissues around Agrobacterium tumefaciens-infiltrated sites on CsWRKY11- and/or CsNPR1-overexpressed cotyledons following NBT staining, in the presence or absence of SA. E Relative transcript levels of CsNYE1 in CsWRKY11- and/or CsNPR1-overexpressed cotyledons, in the presence or absence of SA. F Relative transcript levels of CsRBOHB in CsWRKY11- and/or CsNPR1-overexpressed samples, in the presence or absence of SA. In (A-F), 300 μM SA was used for treatment, cotyledons infiltrated with empty vector (pCHF3) transfected-Agrobacterium tumefaciens were used as controls. Cotyledon samples were photographed and then harvested for analysis four days post treatment. G ChIP-qPCR assay of the binding of CsWRKY11 to CsNYE1 and CsRBOHB promoters. p35S::CsWRKY11-2 × FLAG- and p35S::2 × FLAG-transfected cucumber cotyledons were used for analysis. Enrichment of the target fragments was normalized to CsACTIN, the coding regions were used as internal controls. H EMSA of the interaction between CsWRKY11 and W-boxes on CsNYE1 and CsRBOHB promoters. Promoter fragments containing W-boxes were biotin-labeled as probes, with the same fragments unlabeled being used as competitors. I Dual-luciferase analysis of the effects of CsWRKY11 and CsNPR1 on the activity of pCsNYE1::FfLUC and pCsRBOHB::FfLUC. pCsNYE1::FfLUC or pCsRBOHB::FfLUC was co-introduced into tobacco leaves with p35S::CsWRKY11 or p35S::CsNPR1 via the mediation of Agrobacterium tumefaciens, leaves transfected with Agrobacterium tumefaciens containing empty vectors were used as controls, SA treatment was implemented by spraying 300 μM SA solution onto the surface of tobacco leaves. Data are means ± SD (n = 3 biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001 (t-test), and different letters indicate significant difference at P < 0.05 (one-way ANOVA test) |
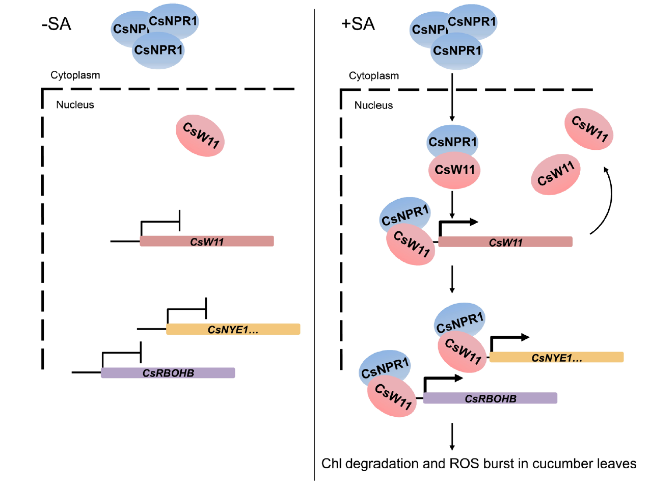
Discussion
Fig. 7 A proposed model of CsNPR1 and CsWRKY11 involved in synergistically regulating SA-triggered chlorophyll degradation and ROS production. In present of SA, CsNPR1 is depolymerized to monomer to enter the nucleus. Nucleus-located CsNPR1 rapidly recruits the transcription factor CsWRKY11 to bind to the W-box on the promoter of CsWRKY11; after amplifying the primary SA signal via self-activation, CsWRKY11 further effects synergistically with CsNPR1 to up-regulate the expression of chlorophyll degradation and ROS biosynthesis related genes, thereby leading to SA-triggered leaf de-greening and ROS burst in cucumber |