Core
Gene and accession numbers
Introduction
Results
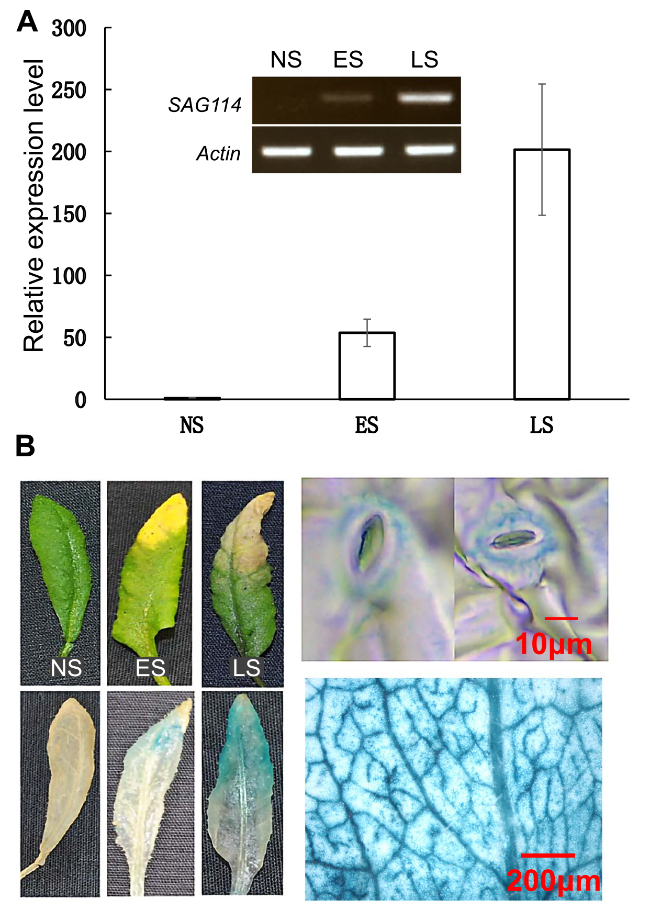
SAG114 SnRK3.25 was specifically expressed in senescing leaves
Fig. 1 The leaf senescence-specific expression of SAG114 SnRK3.25 in Arabidopsis. A Relative expression levels of SAG114 in leaves at different stage in Arabidopsis revealed by qPCR analysis. NS, fully expanded non-senescent leaves; ES, early senescence with up to 25% leaf yellowing; LS, late senescence with > 50% leaf yellowing. The data are presented as the means ± SE (n ≥ 6). Insert represents the semi-quantitative PCR product of the SAG114 transcripts with 28 cycles. B GUS staining of leaves of the PSAG114-GUS transgenic plants. The two panels on the right are close-ups of the GUS stained leaves and stomata |
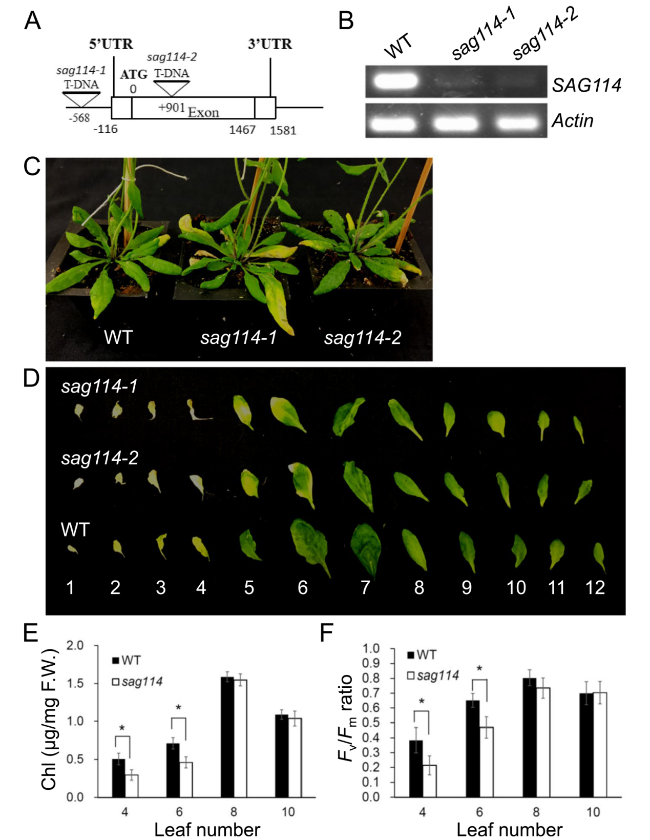
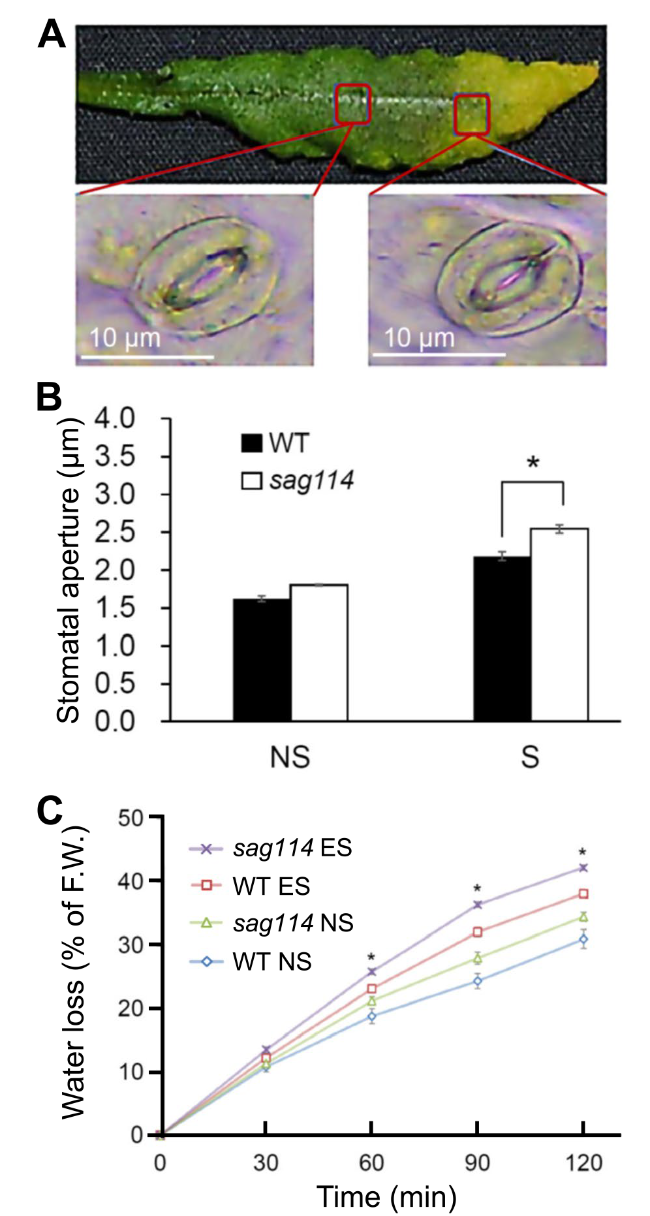
The sag114 null mutants exhibited early leaf senescence and fast water loss phenotypes
Fig. 2 Molecular and functional analyses of SAG114 SnRK3.25 in leaf senescence. A Diagram of SAG114 gene structure and T-DNA insertion sites. There was no intron in the gene. The Arabidopsis T-DNA line SALK_060162 was designated as sag114-1, and SALK_079011 as sag114-2. B RT-PCR analysis of the SAG114 expression in senescing leaves of wild-type (WT), sag114-1 and sag114-2 mutant plants. C Phenotypes of age-matched WT, sag114-1 and sag114-2 plants. D Alignment of age-matched rosette leaves detached from the respective plants (the leaves were counted from bottom). Both sag114-1 and sag114-2 null mutants showed almost the same early leaf senescence phenotype, and only sag114 designation will be used for hereafter analyses. The chlorophyll (Chl) contents (E) and the Fv/Fm ratios (F) in leaves of WT and two sag114 null mutants. Mean values of four samples ± se are shown (n ≥ 3). Asterisks indicate significant differences between wild-type and transgenic plants (Student’s t test, P < 0.05) |
Fig. 3 Larger stomatal aperture and faster water loss in leaves of sag114 null mutant compared with WT. A Example of a senescing leaf of sag114 showing a pair of guard cells with larger stomatal aperture in senescent part and smaller aperture in non-senescent part of the leaf. B Significantly larger aperture in senescent leaves of sag114 than WT. NS, non-senescent leaves without any yellowing; S, senescent leaves that are fully yellowed. C Faster water loss in sag114. Asterisks indicate significant differences between WT and sag114 plants (Student’s t test, P < 0.05) |
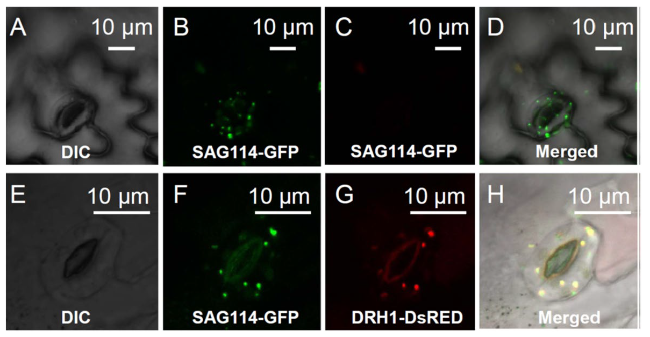
SAG114 was localized in Golgi apparatus
Fig. 4 Localization of SAG114 SnRK3.25 in the Golgi apparatus. A Differential interference contrast (DIC) image of the epidermis of a transgenic plant expressing GFP under the direction of SAG114 promoter (as a control). B The GFP expression in A imaged using the eGFP channel setting of Leica DM5500. C No GFP signal shown in B could be imaged using the DsRED channel setting of Leica DM5500. D Merged image of A-C showing that the SAG114-GFP fusion protein localized to the Golgi apparatus and/or mitochondria. E Differential interference contrast (DIC) image of a senescing leaf epidermis of a transgenic plant containing GFP-tagged SAG114 (SAG114-GFP) and a Golgi marker DsRFP-tagged ERH1 (ERH1-DsRED). F The GFP expression in the guard cells shown in E imaged using the eGFP channel setting of a confocal microscope (Leica DM5500). G Red fluorescent protein expression in the guard cells shown in E taken using the DsRED channel setting of the confocal microscope (Leica DM5500). H Merged image of E-G showing that the SAG114-GFP fusion protein co-localized with the cis-Golgi marker ERH1-DsRED |
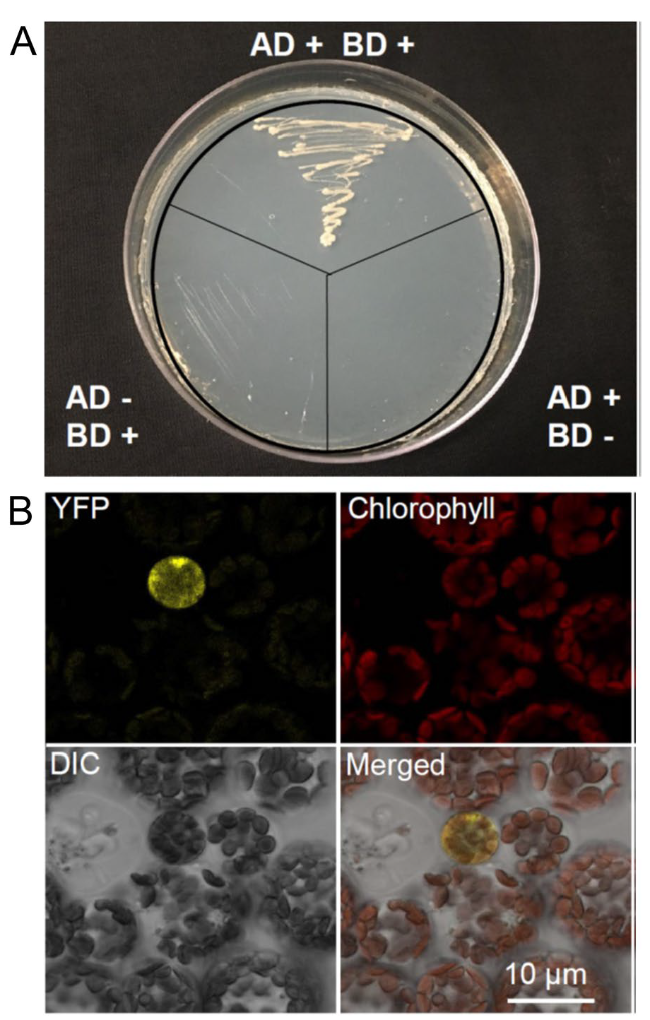
SAG114 physically interacted with SAG113 PP2C in yeast cells and Arabidopsis mesophyll protoplasts
Fig. 5 Physical interactions between SAG113 PP2C and SAG114 SnRK3.25 in both yeast and Arabidopsis leaf cells. A Yeast two-hybrid assay showing the interaction between SAG113 and SAG114.The coding sequences of SAG113 PP2C was fused with the GAL4 activation domain sequence of pGAD424 and the plasmid with the fusion was then transferred into PJ69-4α yeast cells. And the coding sequence of SAG114 SnRK3.25 was fused with the GAL4 binding domain of pGBT9 and the vector was then transferred into PJ69-4A yeast cells. the diploid cells generated via yeast mating were then streaked on SD/-Trp/-Leu/-His/-Ade plate. AD + , yeast cell containing pGAD424-SAG113 only; BD + , yeast cell harboring pGBT9-SAG114 only; AD-, yeast cell without pGAD424-SAG113; BD-, yeast cell without pGBT9-SAG114. B BiFC analysis of the interaction between SAG113-YFPN and SAG114-YFPC in the mesophyll protoplast of Arabidopsis. YFPN and YFPC represent N- and C-terminal half of the yellow fluorescent protein, respectively. The YFP panel, the yellow fluorescence imaged using the eYFP channel setting of Leica DM5500; Chlorophyll, chlorophyll autofluorescence; DIC, differential interference contrast image of the leaf mesophyll protoplasts. Merged, merged image of above images |
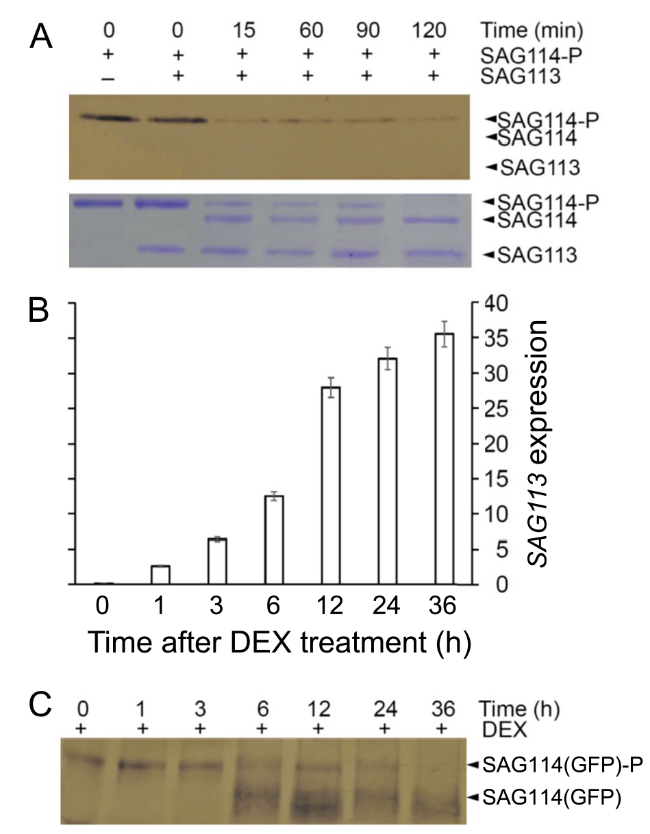
SAG114 was dephosphorylated by SAG113 PP2C in vitro and in planta
Fig. 6 Dephosphorylation of SAG114 SnRK3.25 by SAG113 PP2C in vitro and in planta. A In vitro dephosphorylation assay. SAG113 and the phosphorylated SAG114 (SAG114-P) were co-incubated, separated on SDS-PAGE gel and stained with Coomassie Brilliant Blue (lower panel). The Phospho-Serine/Threonine-specific antibody was used to detect SAG114-P (upper panel). B qRT-PCR analysis of DEX-induced SAG113 expression in non-senescing leaves of Arabidopsis that was constitutively expressing the SAG114-GFP fusion protein [SAG114(GFP)]. C Dephosphorylation of SAG114(GFP)-P by DEX-induced SAG113 in planta. Protein samples from leaves harvested at different time after DEX induction were separated on SDS gel, and the antibody against GFP was used to detect both SAG114(GFP)-P and its dephosphorylated form SAG114(GFP) (as well as its degraded form) |
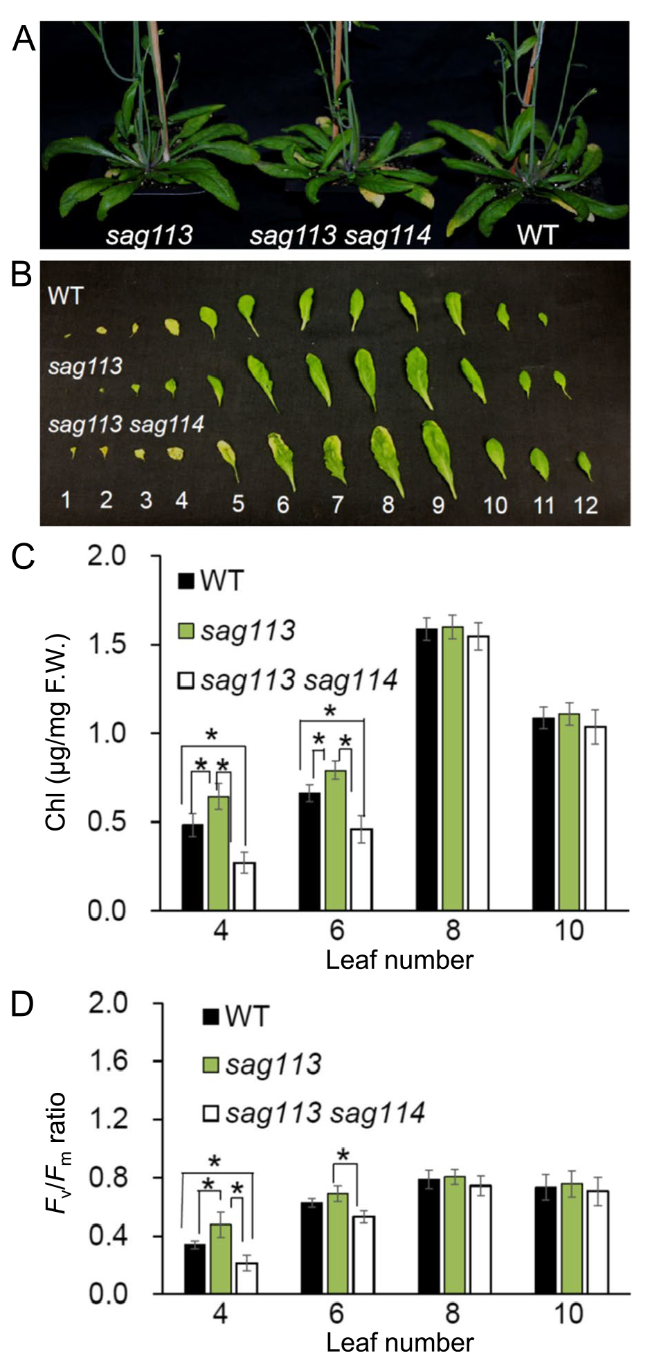
SAG114 was epistatic to SAG113 PP2C
Fig. 7 Epistasis of SAG114 SnRK3.25 to SAG113 PP2C. A Phenotype of age-matched sag113, sag113 sag114 double mutant and WT. B Individual rosette leaves detached from the age-matched plants shown in A. The leaves were counted from bottom. The chlorophyll (Chl) contents (C) and the Fv/Fm ratios (D) in selected leaves shown in B. Mean values of four samples ± se are shown. Asterisks indicate significant differences between indicated plants (Student’s t test, P < 0.05) |
Discussion
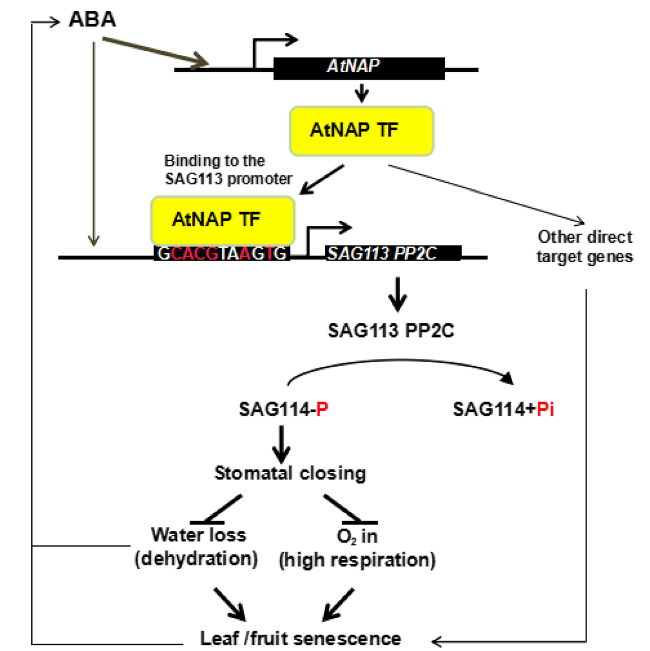
Fig. 8 A working model of SAG114 SnRK3.25. SAG114 SnRK3.25 is the direct target of the ABA-AtNAP-SAG113 PP2C regulatory module (Zhang and Gan 2012). The phosphorylated SAG114 will promote stomatal closure, and the dephosphorylation by SAG113 PP2C will render SAG114 inactive. The figure is modified from Zhang and Gan (2012) |