Core
Gene & Accession Numbers
Introduction
Results
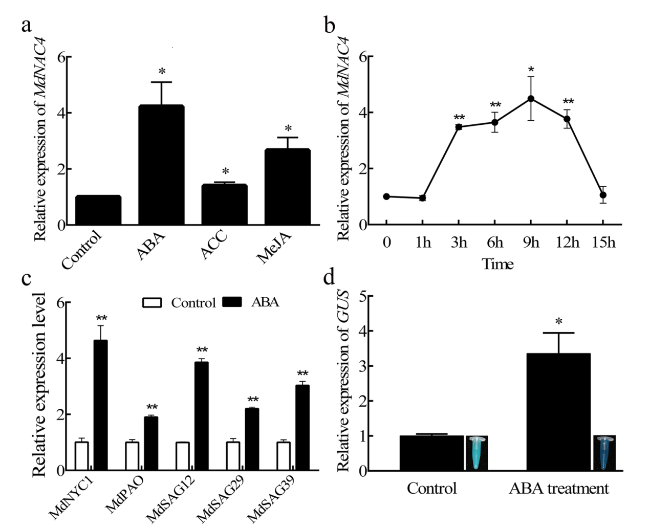
MdNAC4 responds to ABA treatment
Fig. 1 Response of MdNAC4 to ABA treatment. a Expression levels of MdNAC4 in apple seedling leaves treated with different phytohormones. Apple seedlings grown in the nutrient bowl were treated with 50 μM abscisic acid (ABA), 50 μM methyl jasmonate (MeJA), and 50 μM 1-aminocyclopropane-1-carboxylic acid (ACC) for 9 h by adding solutions containing ABA, ACC, and MEJA to the nutrient bowl. b Expression patterns of MdNAC4 after 50 μM ABA treatment. Samples were taken at designated times (0, 1, 3, 6, 9, 12 and 15 h) for RNA extraction. c Expression levels of senescence-related genes (MdNYC1, MdPAO, MdSAG12, MdSAG29 and MdSAG39) after 50 μM ABA treatment for 9 h. (d) GUS staining and qRT-PCR analysis of MdNAC4 promoter transgenic apple calli. Control: no treatment; ABA treatment: 50 μM ABA treatment for 9 h. The expression level in untreated samples was set at 1. Error bars indicate the SDs of the three technical replicates and three biological replicates. Asterisks indicate significant differences between two independent samples according to t tests (*, P < 0.05 and **, P < 0.01) |
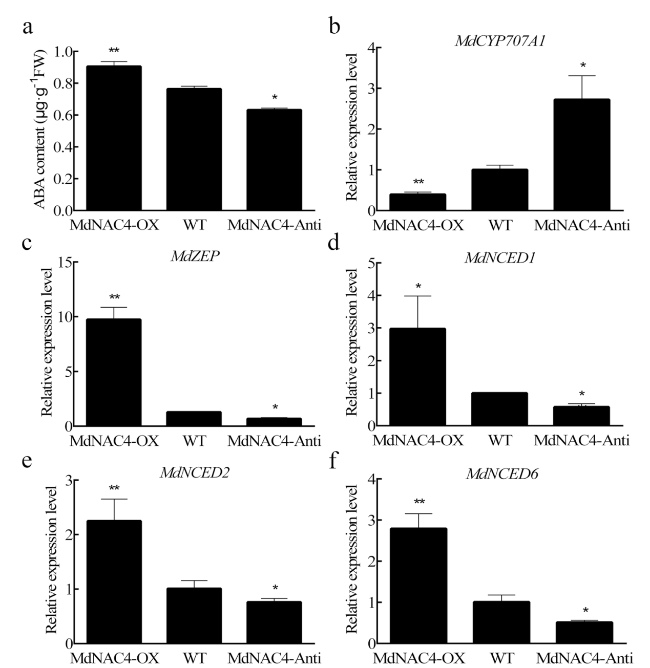
MdNAC4 increases ABA content by regulating the expression of ABA metabolic pathway genes
Fig. 2 ABA content and expression levels of ABA metabolism-related genes in 2-week-old apple calli. a ABA content. b-f Expression levels of ABA metabolism-related genes. WT: wild-type; MdNAC4-OX: with MdNAC4 overexpression vector; MdNAC4-Anti: with MdNAC4 antisense vector. The WT expression level was set at 1. Error bars indicate the SDs of the three technical replicates and three biological replicates. Asterisks indicate significant differences between two independent samples according to t tests (*, P < 0.05 and **, P < 0.01) |
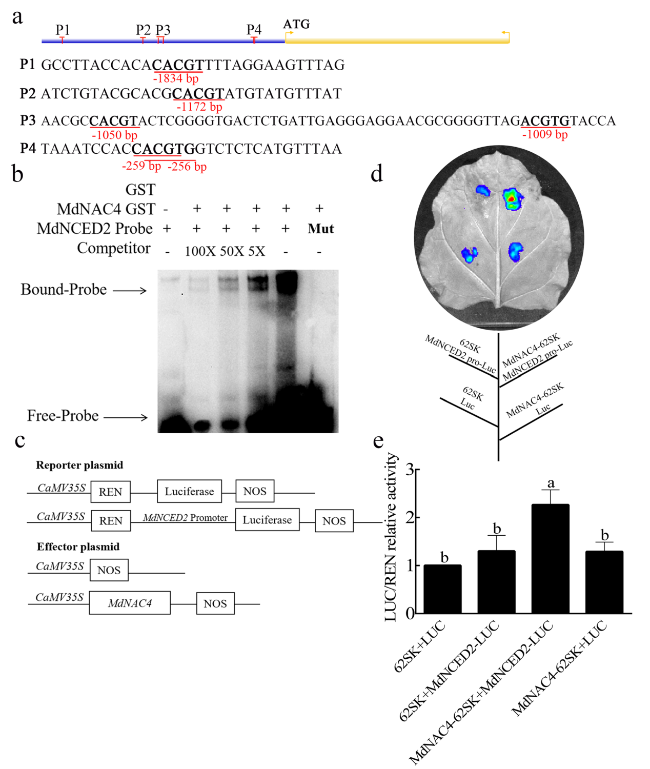
MdNAC4 upregulates the expression of MdNCED2
Fig. 3 MdNAC4 activates the expression of MdNCED2. a Diagram of the MdNCED2 gene promoter region. P1-P4 represent the potential sites to which MdNAC4 might bind. b The electrophoretic mobility shift assay (EMSA) showed that the MdNAC4-GST fusion protein was bound to the MdNCED2 promoter. 5x, 50x and 100x represent the competitor concentrations. The unlabeled probes were used as competitors, with “Mut” representing the mutated probe in which the 5′-ACGTG-3′ motif was replaced by 5′-CCGTC-3′. c Structures of the reporter and effector vectors used in the dual-luciferase assays. The promoter fragment of MdNCED2 was fused into the pGreenII 0800-LUC vector to obtain the reporter plasmid. The MdNAC4 gene was fused to the pGreenII 62-SK vector to generate the effector plasmid. d Dual luciferase assays of tobacco leaves showed that MdNAC4 activated the expression of MdNCED2. e Relative LUC/REN activity analysis verified that MdNAC4 activated the expression of MdNCED2. Tobacco injected with empty vector was used as the control. Error bars indicate the SDs of the three technical replicates and three biological replicates. Different letters above the bars indicate significant differences according to one-way ANOVA (P < 0.05) |
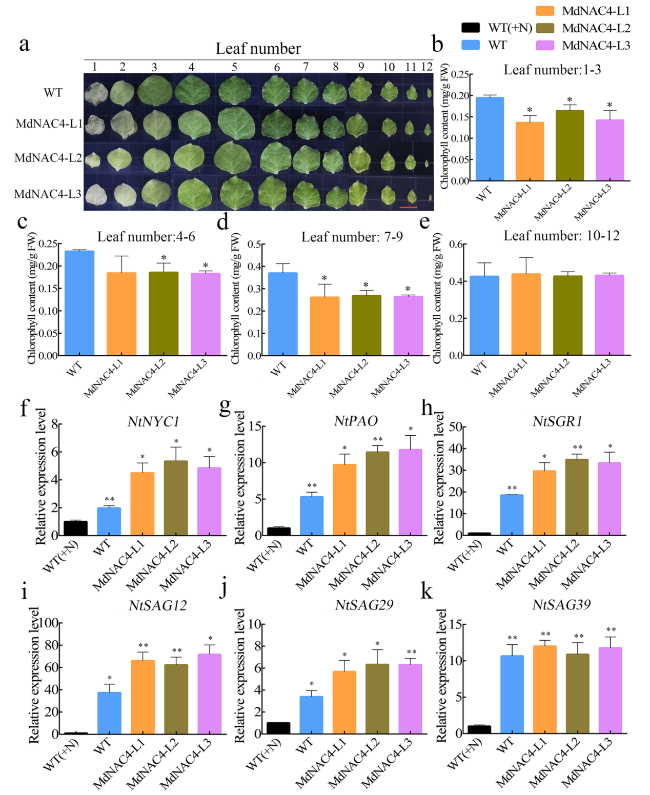
Overexpression of MdNAC4 promotes N deficiency-induced senescence in tobacco leaves
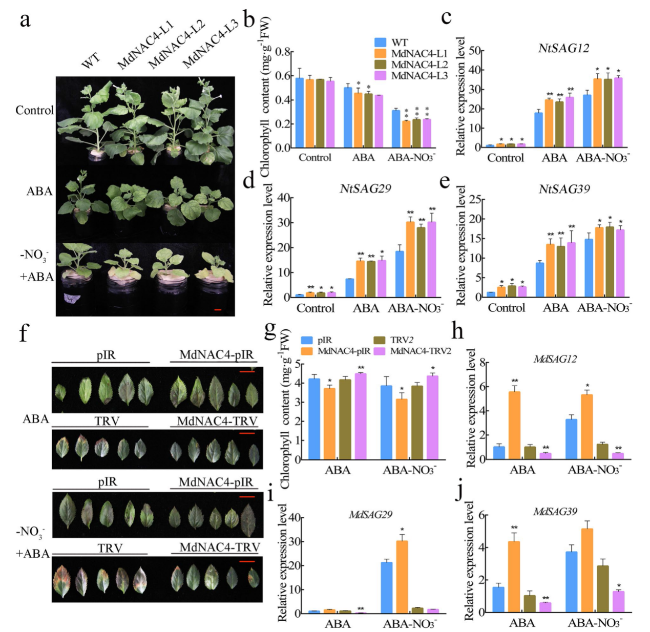
Fig. 4 MdNAC4 caused early senescence induced by N deficiency in tobacco. a Leaf phenotypes of 4-week-old wild-type (Control) and transgenic tobacco (MdNAC4-L1, MdNAC4-L2 and MdNAC4-L3) grown in nitrate-deficient Hoagland nutrient solution for 3 weeks. Tobacco seedlings grown in Hoagland nutrient solution supplied with nitrate before treatment. Representative photographs were taken, where 1-12 represent the numbered leaf positions from the base to tip of tobacco leaves. b-e Total chlorophyll contents of tobacco leaves numbered 1-3, 4-6, 7-9, and 10-16 from 12 plants per indicated genotype. f-k Expression levels of NtNYC1, NtPAO, NtSGR1, NtSAG12, NtSAG29 and NtSAG39 in N-deficient wild-type and MdNAC4-overexpressing tobacco plants for 3 weeks. The expression level in the WT supplied with nitrate (+N) was set at 1. Error bars indicate the SDs of the three technical replicates and three biological replicates. Asterisks indicate significant differences between two independent samples according to t tests (*, P < 0.05 and **, P < 0.01) |
MdNAC4 positively regulates the expression of senescence-related genes
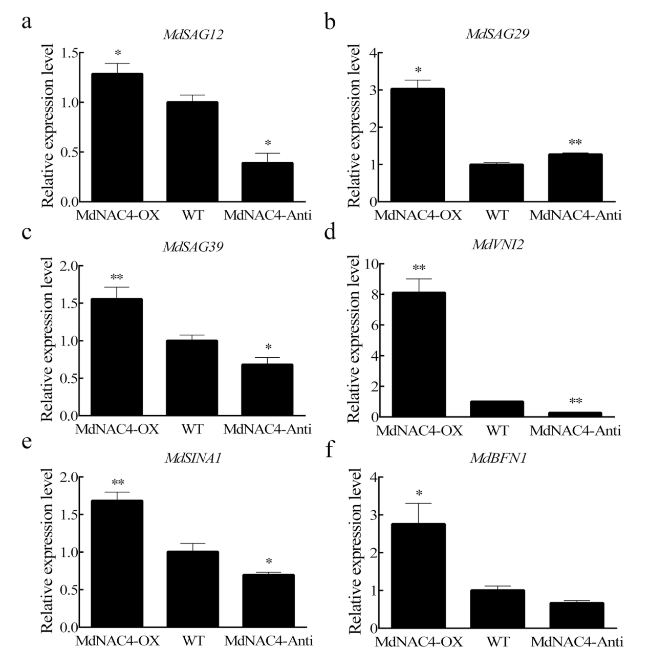
Fig. 5 Expression levels of senescence-related genes (a-f MdSAG12, MdSAG29, MdSAG39, MdVNI2, MdSINA1, and MdBFN1) in 2-week-old apple calli. WT: wild-type; MdNAC4-OX: with MdNAC4 overexpression vector; MdNAC4-Anti: with MdNAC4 antisense vector. The WT expression level was set at 1. Error bars indicate the SDs of the three technical replicates and three biological replicates. Asterisks indicate significant differences between two independent samples according to t tests (*, P < 0.05 and **, P < 0.01) |
MdNAC4 upregulates the expression of MdSAG39
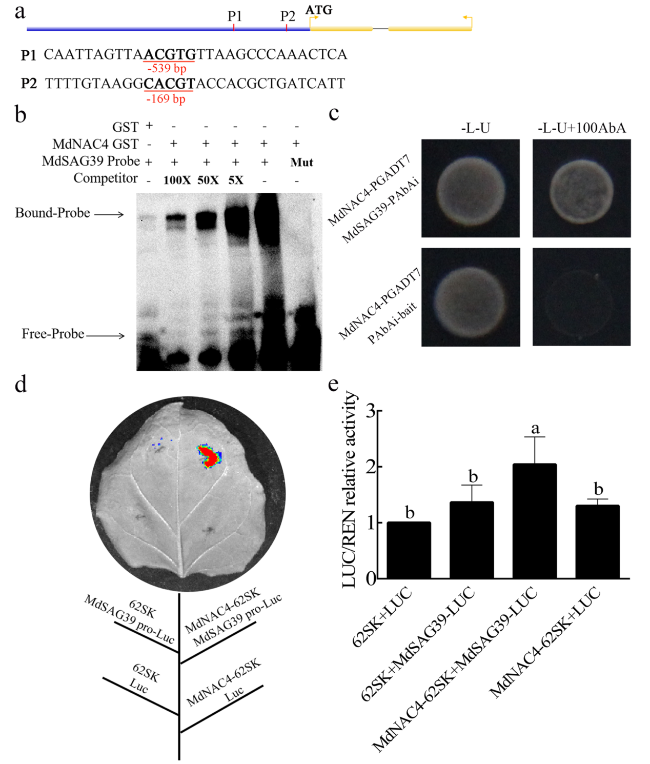
Fig. 6 MdNAC4 activates the expression of MdSAG39. a Diagram of the MdSAG39 gene promoter region. P1 and P2 represent the potential sites to which MdNAC4 might bind. b The electrophoretic mobility shift assay (EMSA) showed that the MdNAC4-GST fusion protein bound to the MdSAG39 promoter. 5x, 50x and 100x represent the competitor concentrations. Unlabeled probes were used as competitors, with “Mut” representing the mutated probe in which the 5′-ACGTG-3′ motif was replaced by 5′-CCGTC − 3′. c A yeast one-hybrid (Y1H) assay revealed the interaction between MdNAC4 and the MdSAG39 promoter. The cotransformed yeast strains were grown on SD/−L-U and SD/−L-U + 100 mM AbA medium for 3 days. d Dual luciferase assays of tobacco leaves showed that MdNAC4 activated the expression of MdSAG39. e Relative LUC/REN activity analysis verified that MdNAC4 activated the expression of MdNCED2. Tobacco injected with the empty vector was used as the control. Error bars indicate the SDs of the three technical replicates and three biological replicates. Different letters above the bars indicate significant differences according to one-way ANOVA (P < 0.05) |
ABA enhances the function of MdNAC4 in leaf senescence induced by N deficiency
Fig. 7 ABA enhances the senescence symptoms of MdNAC4 transgenic tobacco and apple leaves under N deficiency conditions |
The MdNAC4 protein interacts with the MdPYL4 protein
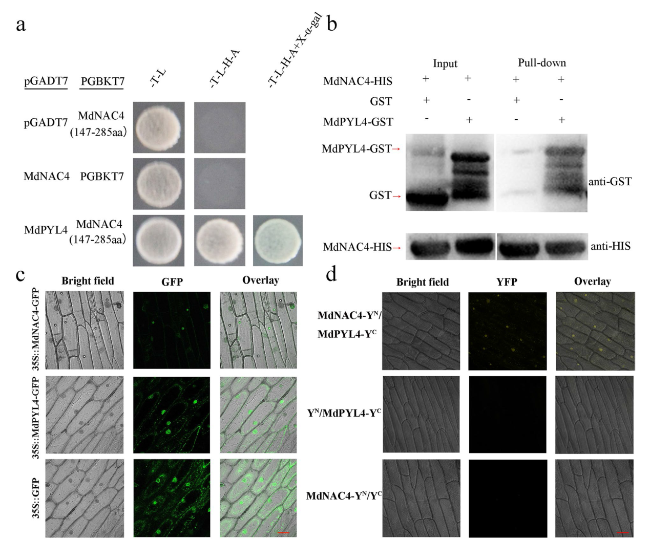
Fig. 8 MdNAC4 interacts with MdPYL4. a Interaction between MdNAC4 and MdPYL4 in the Y2H assay. Full-length MdPYL4 was fused into the pGADT7 vector to obtain a recombinant plasmid (pGADT7-MdPYL4). The recombinant plasmid pGBKT7-MdNAC4 was obtained by fusing the MdNAC4 fragment without the autonomous activation domain into the pGBKT7 vector. Empty pGADT7 + pGBKT7-MdNAC4 (147-285 aa) and empty pGBKT7 + pGADT7-MdNAC4 were used as controls. The cotransformed yeast grown in SD (−T/−L), SD (−T/−L/−H/−A) and SD (−T/−L/−H/−A + X-α-gal) medium are indicated. b Interaction between the MdNAC4 and MdPYL4 proteins in the pull-down assay. The glutathione transferase (GST), MdPYL4-GST and MdNAC4-HIS proteins were induced by isopropyl thiogalactoside in Escherichia coli. The GST and MdPYL4-GST proteins were incubated with the MdNAC4-HIS protein, and the protein mixture was purified using the GST purification kit. c Subcellular localization of the MdPYL4 protein and the MdNAC4 protein. GFP, green fluorescent protein. d BiFC assays showed that the MdNAC4 protein interacted with the MdPYL4 protein. YFP, yellow fluorescent protein |
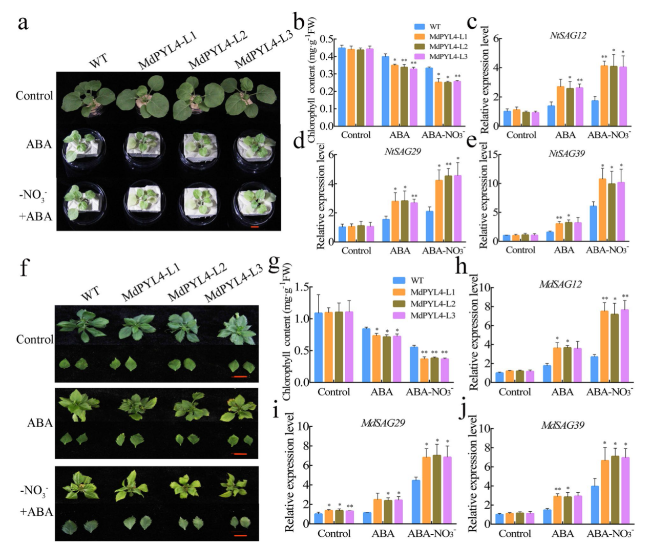
Overexpression of MdPYL4 promotes ABA-induced leaf senescence
Fig. 9 Overexpression of MdPYL4 promotes ABA-induced leaf senescence under N deficiency conditions |
Discussion
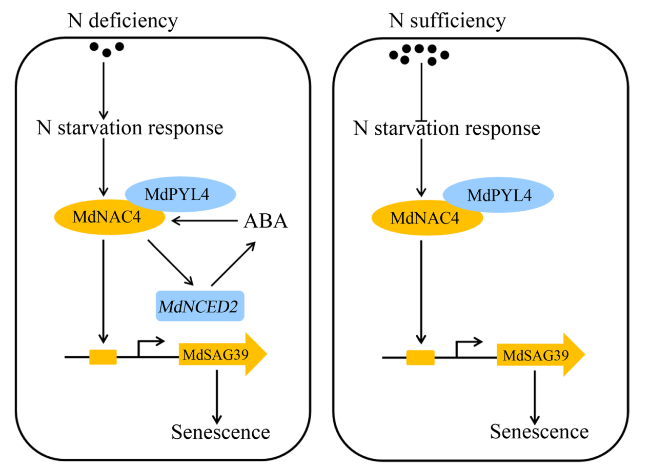
Fig. 10 A working model of the MdNAC4-mediated regulation of N deficiency-induced leaf senescence in apple. Under N-deficient conditions, N starvation-responsive genes are activated, which induces the expression of MdNAC4. MdNAC4 directly activates the expression of the ABA biosynthesis gene MdNCED2 and further promotes ABA biosynthesis. The ABA receptor protein MdPYL4 interacts with the MdNAC4 protein and enhances the response of MdNAC4 to N deficiency. MdNAC4 directly binds to the SAG MdSAG39 and activates its expression, thus promoting leaf senescence induced by N deficiency. Under N-sufficient conditions, N starvation-responsive genes are suppressed. MdPYL4 interacts with MdNAC4 proteins to activate the expression of SAG MdSAG39, thus regulating leaf senescence |