Core
Gene and accession numbers
Introduction
Results
Transcriptional expression analysis of CmBBX7 in chrysanthemum cv. ‘Yuuka’
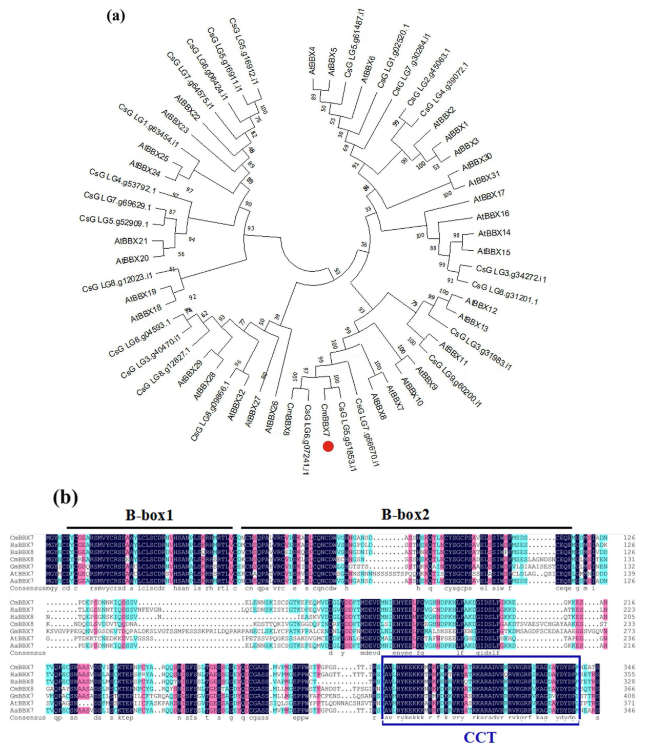
Fig. 1 CmBBX7 encodes a BBX family subgroup II protein. a Phylogenetic relationships of CmBBX7 and BBX sequences from Arabidopsis and Chrysanthemum seticuspe. Protein sequences of the 32 BBXs in Arabidopsis were obtained from The Arabidopsis Information Resource (TAIR) database, the sequence of BBX8 in chrysanthemum was obtained from the National Center for Biotechnology Information (NCBI), and the genome sequence and annotation data of C. seticuspe are available at PlantGarden with Project ID PRJDB7468. The percentage of trees in which the associated taxa clustered together is shown next to the branches. b Alignment of the deduced polypeptide sequences of CmBBX7 with those of other plant BBXs. The B-box1 and B-box2 domains are indicated by the black lines above the sequences. The CCT motif is indicated by the blue lines around the sequences |
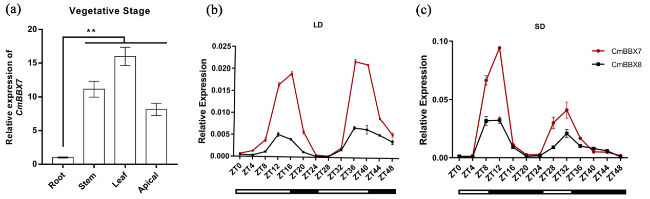
Fig. 2 Transcriptional expression analysis of CmBBX7 in chrysanthemum cv. ‘Yuuka’. a Relative mRNA expression levels of CmBBX7 in various parts of the plant during the vegetative stage. Error bars indicate the standard errors for three biological replicates. Significant differences are indicated with asterisks (**p < 0.01, ANOVA with Tukey’s post-hoc test). b,c The transcriptional response of CmBBX7, and CmBBX8 to varying photoperiods [long day (LD) and short day (SD)]. The abscissa indicates the sampling time point. Values shown are the means (n = 3) and the error bars represent the standard errors of the mean |
CmBBX7 accelerates the floral transition
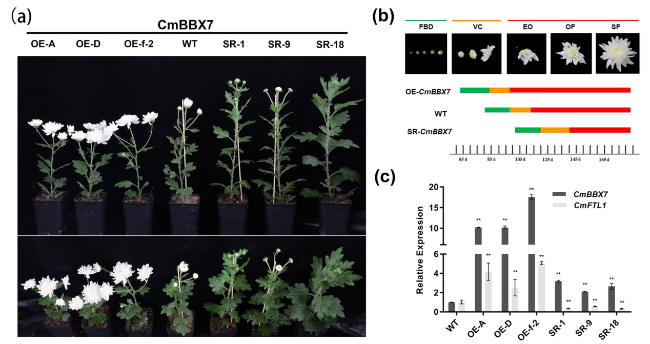
Fig. 3 CmBBX7 accelerates the floral transition. a The phenotypes of the CmBBX7 transgenic lines and wild-type (WT) plants. The upper panel shows the elevation view and the lower panel shows the top view. OE: overexpression lines; SR: transcriptional repression lines. b Statistics of flowering time. FBD: flower bud development stage; VC: visible color stage; EO: early opening stage; OF: opened flower stage; SF: senescing flower stage. The lower bars show the time taken to grow the plants from planting to the various stages of flowering. c Relative expression levels of CmBBX7 and CmFTL1 in transgenic plants. Error bars indicate the standard deviations for three biological replicates. Significant differences are indicated with asterisks (**p < 0.01, ANOVA with Tukey’s post-hoc test) |
CmBBX7 directly regulates CmFTL1
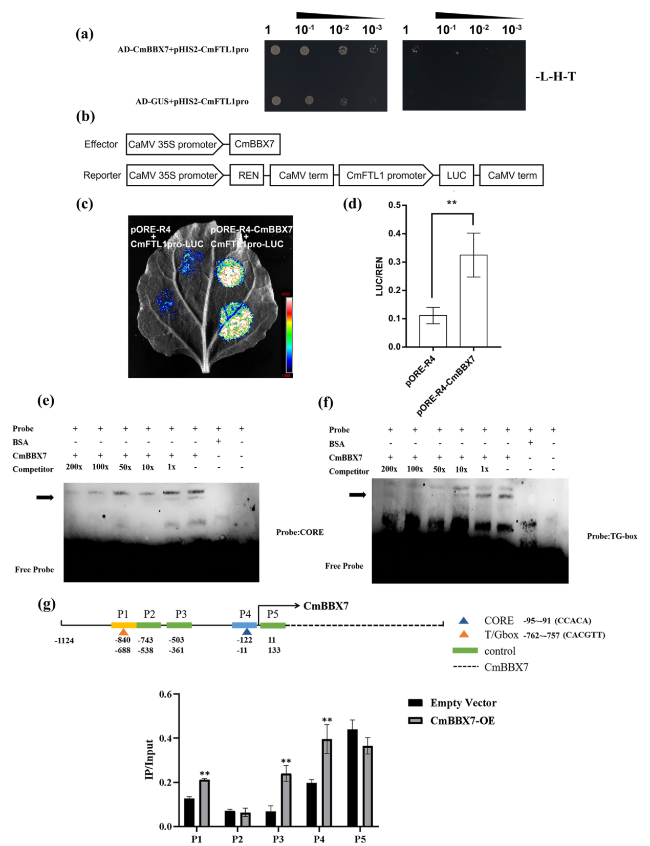
Fig. 4 CmBBX7 regulates CmFTL1 directly. Interactions of CmBBX7 protein with the promoters of CmFTL1 in the yeast one-hybrid assay. SD/-T/-H/-L indicates Trp, His, and Leu synthetic dropout medium, respectively. The 3-AT concentration is 60 mM for CmFTL1pro. b Effector and reporter vector construction diagrams for the dual-luciferase assays. c Transient expression in N. benthamiana leaves of the pCmFTL1::LUC transgene. Graph showing luminescence intensity. d The ratio of LUC to REN activity. Error bars indicate the SD for six biological replicates. Significant differences are indicated by asterisks (** p < 0.01, ANOVA, Tukey’s correction). e-f EMSA of CmBBX7 binding to the CORE and TG-box elements of CmFTL1 promoter. ‘ + ’ indicates presence and ‘ − ’ indicates absence. g ChIP-qPCR of the enrichment of DNA fragments CORE (-95 to -91 bp) and TG-box (-762 to -757 bp) in the CmFTL1 promoter. Error bars indicate the SD for six biological replicates. Significant differences are indicated by asterisks (** p < 0.01, ANOVA, Tukey’s correction). |
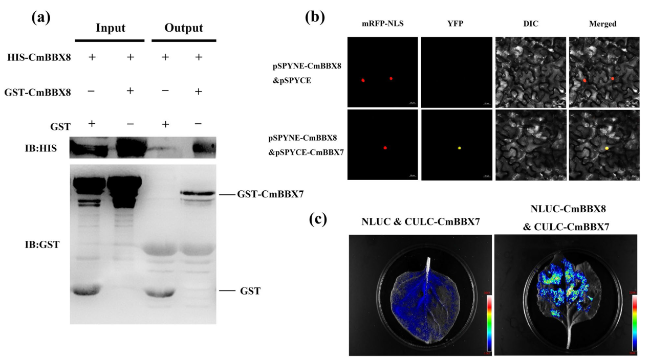
CmBBX7 interacts with CmBBX8
Fig. 5 CmBBX7 interacts with CmBBX8. a Pull-down assays of the interaction between CmBBX7 and CmBBX8. The arrows show the target protein sizes. GST-empty vector and His-CmBBX8 were used as controls. b BiFC assay. mRFP-NLS: images taken in the red fluorescence channel; YFP: images taken in the yellow fluorescence channel; DIC: images taken in bright light; Merged: both overlay plots; bars = 20 μm. c Firefly luciferase complementation imaging (LCI) assay verified the interaction between CmBBX7 and CmBBX8 in N. benthamiana leaves |
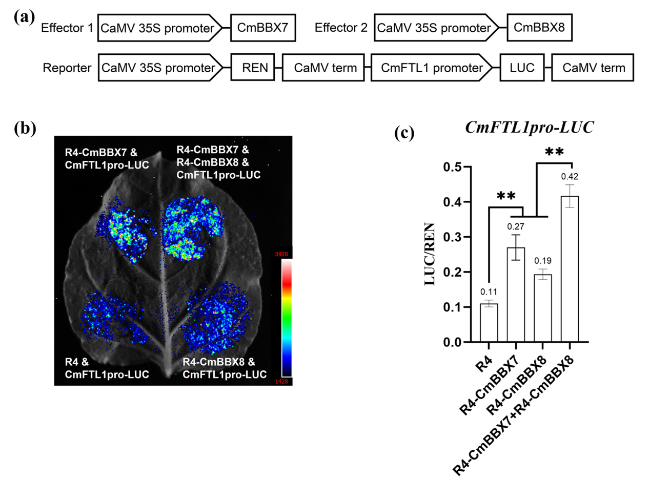
CmBBX7 and CmBBX8 synergistically regulate CmFTL1 expression
Fig. 6 CmBBX7 and CmBBX8 synergistically regulate CmFTL1 expression. a Effector and reporter vector construction diagrams for dual-luciferase assays. b Transient expression in N. benthamiana leaves of the pCmFTL1::LUC transgene. The graph shows the luminescence intensity. c The ratio of LUC to REN activity. Error bars indicate the standard deviations for six biological replicates. Significant differences are indicated with asterisks (**p < 0.01, ANOVA with Tukey’s post-hoc test) |
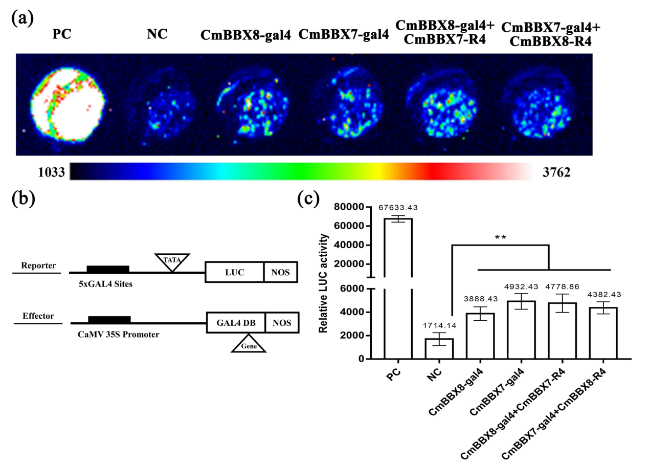
Fig. 7 CmBBX7 and CmBBX8 do not mutually affect each other’s activity. Fluorescence images: from blue to red, the fluorescence value increases gradually. PC: positive control (AtARF5); NC: negative control (empty vector). b Vector construction diagrams for the reporter and effector constructs used in the transcription activity assay. The reporter construct contains five copies in tandem of the GAL4-binding sites upstream of the promoter (TATA), the firefly gene for luciferase, and Nos. The effector constructs contain the GAL4 DNA-binding domain (GAL4 DB) between the CaMV 35S promoter and Nos. c Relative LUC activity. Error bars indicate the standard deviations for six biological replicates. Significant differences are indicated with asterisks (**p < 0.01, ANOVA with Tukey’s post-hoc test) |