

Journal of Surgery Concepts & Practice ›› 2024, Vol. 29 ›› Issue (05): 414-425.doi: 10.16139/j.1007-9610.2024.05.09
• Original article • Previous Articles Next Articles
CHU Yudan, SUN Haidong, CUI Ran, ZHENG Hong( )
)
Received:2024-08-25
Online:2024-09-25
Published:2025-01-23
Contact:
ZHENG Hong
E-mail:zzhzhzjy@163.com
CLC Number:
CHU Yudan, SUN Haidong, CUI Ran, ZHENG Hong. Expression and clinical significance of disulfidptosis-related LncRNA in gastric cancer[J]. Journal of Surgery Concepts & Practice, 2024, 29(05): 414-425.
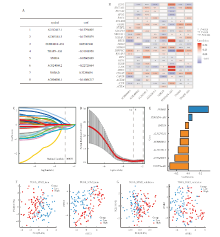
Fig 2
Construction of risk score model based on DRLncs in gastric cancer (DRG-RS) A: The top 8 prognostic DRLncs; B: Heat map showing the correlation between DRGs and LncRNAs; C: Path plots of the LASSO regression coefficients; D: Cross-validation curves of DRLs in the LASSO regression; E: Regression coefficient of Signature; F: PCA and t-SNE were used to analyze the distribution of high and low risk groups in the training set; G: PCA and t-SNE analysis of the validation set for the distribution of high and low risk groups.

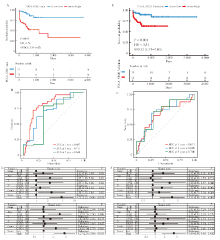
Fig 3
Evaluation and validation of the independent prognostic ability of DRG-RS for gastric cancer A: The Kaplan-Meier (K-M) survival analysis of gastric cancer patients between high- and low-risk DRG-RS groups in the training set; B: The receiver ROC curves showed the predictive accuracy in 1-, 3-, and 5-year of the predictive risk model in the training set; C: The Kaplan-Meier (K-M) survival analysis of gastric cancer patients between high- and low-risk DRG-RS groups in the validation set; D: The ROC curves showed the predictive accuracy in 1-, 3-, and 5-year of the predictive risk model in the validation set; E: Univariate COX analysis and multivariate COX analysis of the training set; F: Univariate COX analysis and multivariate COX analysis of the validation set.


Fig 4
Construction of a nomogram combining clinical characteristics. A: The nomogram to predict the 1-, 3-, and 5-year overall survival (OS) rate of gastric cancer patients; B:) The calibration curve for evaluating the accuracy of the nomogram model in 1-, 3-, and 5-year categories; C: The time C-index curve of the Nomogram model; D: ROC curve of 1-year, 3-year and 5-year prognosis; E: ROC curve of the predictive power of the Nomogram model and clinicopathological parameters.
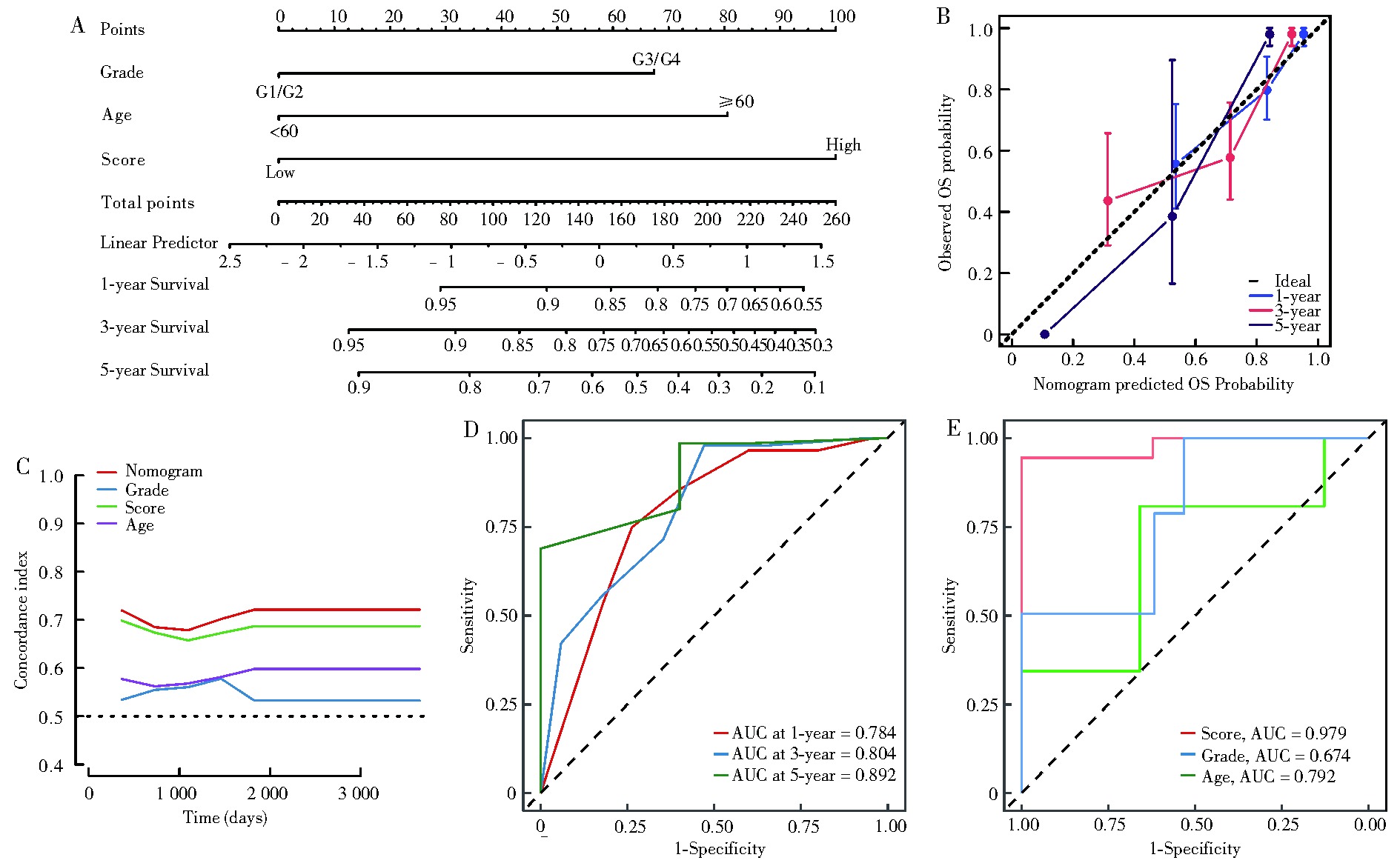
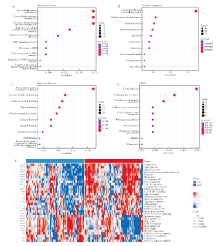
Fig 5
Analysis of DRG-RS related function A-C: GO enrichment analysis based on differential genes showed biological process (A: BP), cell component (B: CC) and molecular function (C: MF), respectively; D: KEGG pathway analysis based on differential genes; E: Heat map of tumor characteristic functional pathway activity.
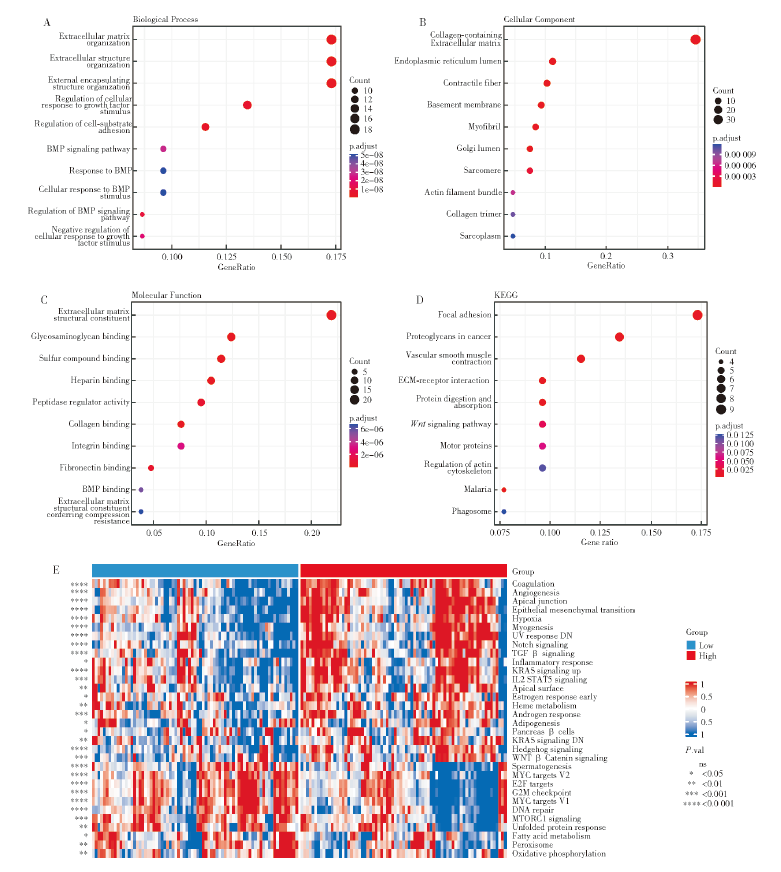
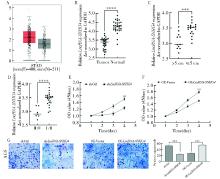
Fig 8
LncRNA-SNHG4 promotes the proliferation and migration of gastric cancer cells A: The expression of LncRNA-SNHG4 in gastric cancer tissues was analyzed based on GEPIA; B: The expression of LncRNA-SNHG4 in gastric cancer tissues was analyzed based on gastric cancer samples; C: The correlation analysis between the expression of LncRNA-SNHG4 and tumor size in gastric cancer tissues; D: The correlation analysis between the expression of LncRNA-SNHG4 and tumor stage in gastric cancer tissues; E: Knockdown of LncRNA-SNHG4 in AGS cells inhibited the proliferation of gastric cancer cell; F:Overexpression of LncRNA-SNHG4 in AGS cells promoted the proliferation of gastric cancer cell; G: Knockdown of LncRNA-SNHG4 in AGS cells inhibited the migration of gastric cancer cells, while LncRNA-SNHG4 overexpression promoted the migration of gastric cancer cells.
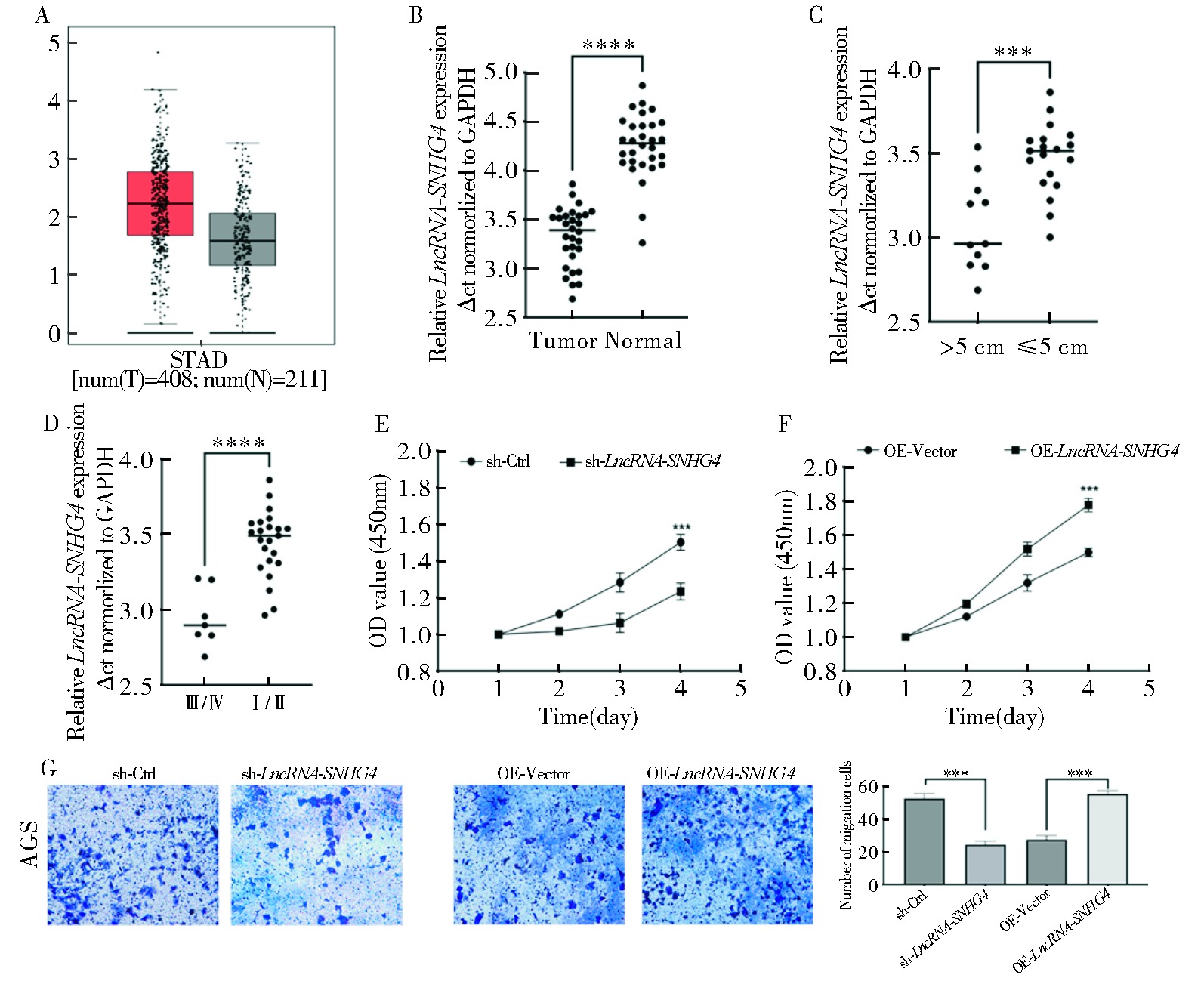
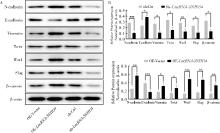
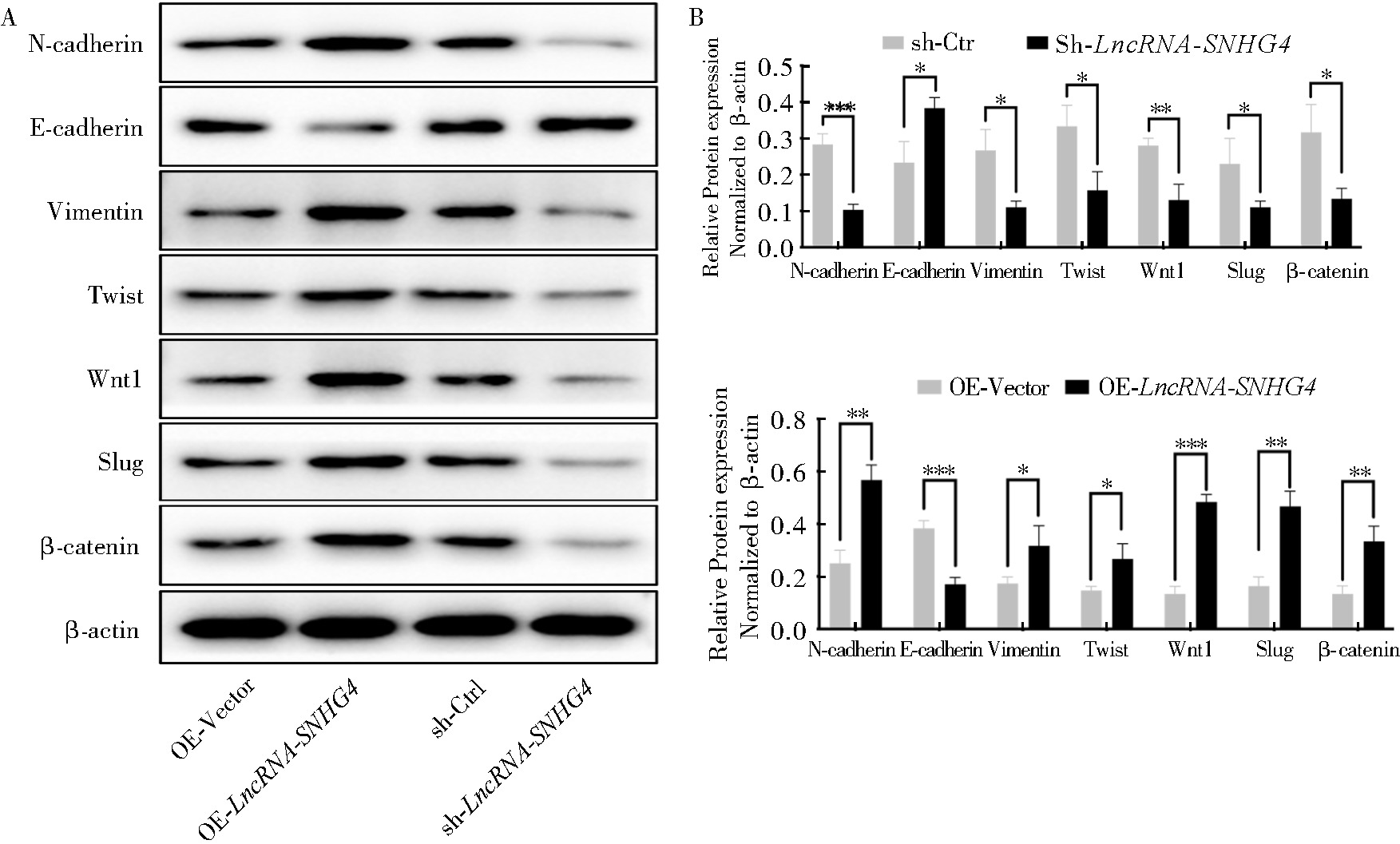
Fig 9
LncRNA-SNHG4 promotes the activation of Wnt signaling pathway A:The expression of proteins associated with Wnt signaling pathway detected by Western Blotting in different groups; B: Statistical plot showed expression of the proteins associated with Wnt signaling pathway in different groups.
| [1] | TORRE L A, BRAY F, SIEGEL R L, et al. Global cancer statistics,2012[J]. CA Cancer J Clin, 2015, 65(2):87-108. |
| [2] | GUAN W L, HE Y, XU R H. Gastric cancer treatment: recent progress and future perspectives[J]. J Hematol Oncol, 2023, 16(1):57. |
| [3] |
WEI L, SUN J, ZHANG N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance[J]. Mol Cancer, 2020, 19(1):62.
doi: 10.1186/s12943-020-01185-7 pmid: 32192494 |
| [4] | LE J, PAN G, ZHANG C, et al. Targeting ferroptosis in gastric cancer: strategies and opportunities[J]. Immunol Rev, 2024, 321(1):228-245. |
| [5] |
NEWTON K, STRASSER A, KAYAGAKI N, et al. Cell death[J]. Cell, 2024, 187(2):235-256.
doi: 10.1016/j.cell.2023.11.044 pmid: 38242081 |
| [6] |
HANAHAN D. Hallmarks of cancer: new dimensions[J]. Cancer Discov, 2022, 12(1):31-46.
doi: 10.1158/2159-8290.CD-21-1059 pmid: 35022204 |
| [7] |
CARNEIRO B A, EL-DEIRY W S. Targeting apoptosis in cancer therapy[J]. Nat Rev Clin Oncol, 2020, 17(7):395-417.
doi: 10.1038/s41571-020-0341-y pmid: 32203277 |
| [8] | PISTRITTO G, TRISCIUOGLIO D, CECI C, et al. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies[J]. Aging (Albany NY), 2016, 8(4):603-619. |
| [9] |
ZHANG C, LIU X, JIN S, et al. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance[J]. Mol Cancer, 2022, 21(1):47.
doi: 10.1186/s12943-022-01530-y pmid: 35151318 |
| [10] | GAO W, WANG X, ZHOU Y, et al. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy[J]. Signal Transduct Target Ther, 2022, 7(1):196. |
| [11] | FAUBERT B, SOLMONSON A, DEBERARDINIS R J. Metabolic reprogramming and cancer progression[J]. Science, 2020, 368(6487):eaaw5473. |
| [12] |
MAO C, WANG M, ZHUANG L, et al. Metabolic cell death in cancer: ferroptosis, cuproptosis, disulfidptosis, and beyond[J]. Protein Cell, 2024, 15(9):642-660.
doi: 10.1093/procel/pwae003 |
| [13] |
ZHENG P, ZHOU C, DING Y, et al. Disulfidptosis: a new target for metabolic cancer therapy[J]. J Exp Clin Cancer Res, 2023, 42(1):103.
doi: 10.1186/s13046-023-02675-4 pmid: 37101248 |
| [14] |
XIAO F, LI H L, YANG B, et al. Disulfidptosis: a new type of cell death[J]. Apoptosis, 2024, 29(9-10):1309-1329.
doi: 10.1007/s10495-024-01989-8 pmid: 38886311 |
| [15] | BRIDGES M C, DAULAGALA A C, KOURTIDIS A. LNCcation: lncRNA localization and function[J]. J Cell Biol, 2021, 220(2):e202009045. |
| [16] | WU Y, WEN X, XIA Y, et al. LncRNAs and regulated cell death in tumor cells[J]. Front Oncol, 2023,13:1170336. |
| [17] | WANG R, YU X, YE H, et al. LncRNA FAM83H-AS1 inhibits ferroptosis of endometrial cancer by promoting DNMT1-mediated CDO1 promoter hypermethylation[J]. J Biol Chem, 2024, 300(9):107680. |
| [18] | LIU S, ZHENG Y, LI S, et al. Integrative landscape analysis of prognostic model biomarkers and immunogenomics of disulfidptosis-related genes in breast cancer based on LASSO and WGCNA analyses[J]. J Cancer Res Clin Oncol, 2023, 149(18):16851-16867. |
| [19] | XU K, ZHANG Y, YAN Z, et al. Identification of disulfidptosis related subtypes, characterization of tumor microenvironment infiltration, and development of DRG prognostic prediction model in RCC, in which MSH3 is a key gene during disulfidptosis[J]. Front Immunol, 2023,14:1205250. |
| [20] |
CHENG J, MA H, YAN M, et al. THAP9-AS1/miR-133b/SOX4 positive feedback loop facilitates the progression of esophageal squamous cell carcinoma[J]. Cell Death Dis, 2021, 12(4):401.
doi: 10.1038/s41419-021-03690-z pmid: 33854048 |
| [21] | JIA W, ZHANG J, MA F, et al. Long noncoding RNA THAP9-AS1 is induced by Helicobacter pylori and promotes cell growth and migration of gastric cancer[J]. Onco Targets Ther, 2019,12:6653-6663. |
| [22] |
DONG Q, QIU H, PIAO C, et al. LncRNA SNHG4 promotes prostate cancer cell survival and resistance to enzalutamide through a let-7a/RREB1 positive feedback loop and a ceRNA network[J]. J Exp Clin Cancer Res, 2023, 42(1):209.
doi: 10.1186/s13046-023-02774-2 pmid: 37596700 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||